5MYP
 
 | | Structure of apo-TbALDH3 | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL | | Authors: | Zoltner, M, Zhang, N, Horn, D, Field, M.C. | | Deposit date: | 2017-01-27 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of an anti-trypanosomal benzoxaborole requires both host and parasite factors
To Be Published
|
|
7N82
 
 | | NMR Solution structure of Se0862 | | Descriptor: | Biofilm-related protein | | Authors: | Zhang, N, LiWang, A.L. | | Deposit date: | 2021-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2.
Proteins, 89, 2021
|
|
5XME
 
 | | Solution structure of C-terminal domain of TRADD | | Descriptor: | Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2017-05-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Sci Rep, 7, 2017
|
|
5XTA
 
 | | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, VirK protein | | Authors: | Yin, S, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2017-06-18 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila, reveals a novel fold for bacterial VirK proteins
FEBS Lett., 591, 2017
|
|
2Z4D
 
 | |
5HKW
 
 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5HKX
 
 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
5HL0
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
5HKY
 
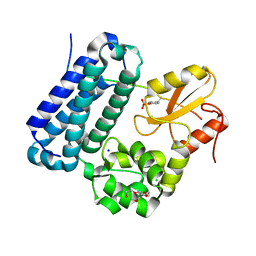 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
7V6V
 
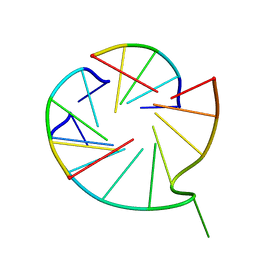 | |
7CSQ
 
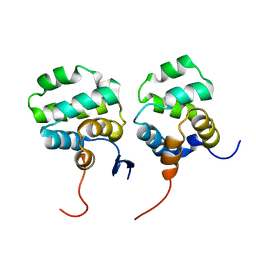 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | Descriptor: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
4MYW
 
 | | Structure of HSV-2 gD bound to nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
5EZK
 
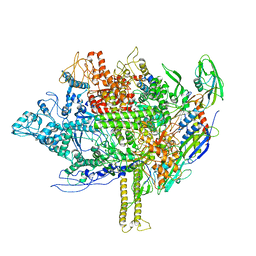 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
4W8F
 
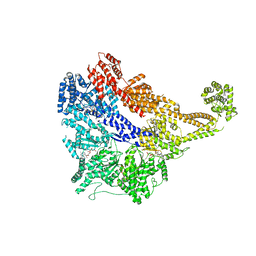 | | Crystal structure of the dynein motor domain in the AMPPNP-bound state | | Descriptor: | Dynein heavy chain lysozyme chimera, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cheng, H.-C, Bhabha, G, Zhang, N, Vale, R.D. | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.541 Å) | | Cite: | Allosteric communication in the Dynein motor domain.
Cell, 159, 2014
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
1WWN
 
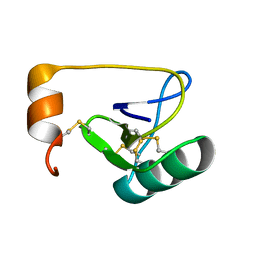 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
4QOS
 
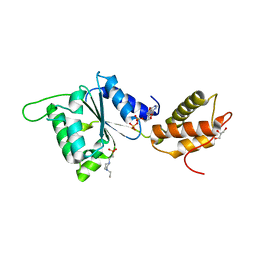 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNR
 
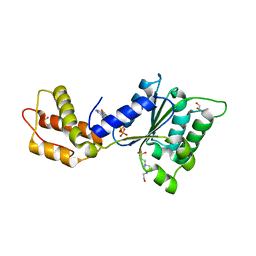 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNM
 
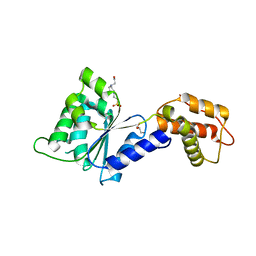 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
1M2S
 
 | | Solution Structure of A New Potassium Channels Blocker from the Venom of Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Toxin BmTX3 | | Authors: | Wang, Y, Li, M, Zhang, N, Wu, G, Hu, G, Wu, H. | | Deposit date: | 2002-06-25 | | Release date: | 2004-04-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of BmTx3B, a member of the scorpion toxin subfamily alpha-KTx 16
Proteins, 58, 2005
|
|
5XMM
 
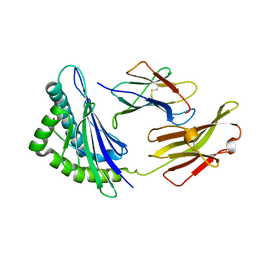 | | FLA-E*01801-167W/S | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5XPV
 
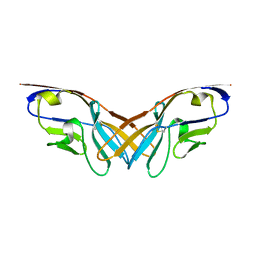 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5XMF
 
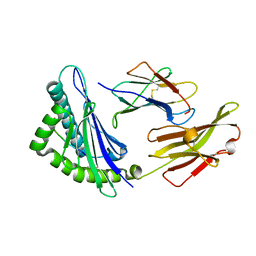 | | Crystal structure of feline MHC class I for 2,1 angstrom | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
