7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
6AFP
 
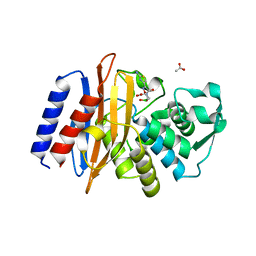 | | Crystal structure of class A b-lactamase, PenL, variant Asn136Asp, from Burkholderia thailandensis, in complex with ceftazidime-like boronic acid | | Descriptor: | ACETATE ION, Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Cao, T.-P, Choi, J.M, Lee, S.H. | | Deposit date: | 2018-08-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Non-catalytic-Region Mutations Conferring Transition of Class A beta-Lactamases Into ESBLs.
Front Mol Biosci, 2020
|
|
6AFM
 
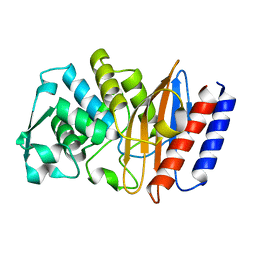 | |
6AFO
 
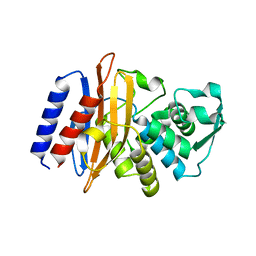 | |
6AFN
 
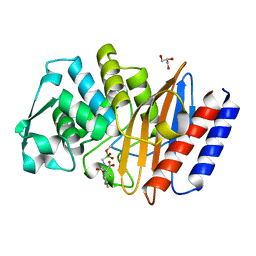 | | Crystal structure of class A b-lactamase, PenL, variant Cys69Tyr, from Burkholderia thailandensis, in complex with ceftazidime-like boronic acid | | Descriptor: | Beta-lactamase, GLYCEROL, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Cao, T.-P, Choi, J.M, Lee, S.H. | | Deposit date: | 2018-08-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-catalytic-Region Mutations Conferring Transition of Class A beta-Lactamases Into ESBLs.
Front Mol Biosci, 2020
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7Y0I
 
 | | Solution structures of ASH1L PHD domain in complex with H3K4me2 peptide | | Descriptor: | ALA-ARG-THR-MLY-GLN-THR-ALA-ARG-LYS-SER-THR-GLY-GLY-LYS-ALA, Histone-lysine N-methyltransferase ASH1L, ZINC ION | | Authors: | Yu, M, Zeng, L. | | Deposit date: | 2022-06-05 | | Release date: | 2022-10-12 | | Method: | SOLUTION NMR | | Cite: | Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer.
Front Oncol, 12, 2022
|
|
