1VYU
 
 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYV
 
 | | beta4 subunit of Ca2+ channel | | Descriptor: | CALCIUM CHANNEL BETA-4SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYT
 
 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
3OGT
 
 | | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3. | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Fraga, R, Mourino, A, Moras, D, Rochel, N. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3.
To be Published
|
|
3CS6
 
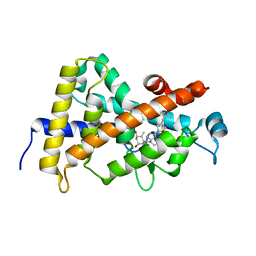 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, P, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
3CS4
 
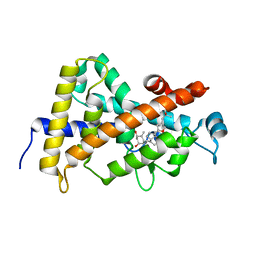 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-9,10-secoandrosta-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, F, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
8Q77
 
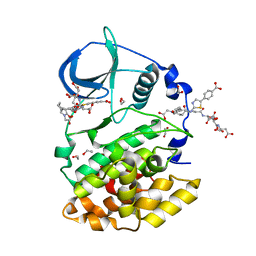 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BISUBSTRATE INHIBITOR ARC-780 | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[12-[[4-[[5-(4-carboxyphenyl)-1,3-thiazol-2-yl]amino]-4-oxidanylidene-butanoyl]-(2-hydroxy-2-oxoethyl)amino]dodecanoylamino]-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-oxidanyl-4-oxidanylidene-butanoyl]amino]butanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.255 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
6SPX
 
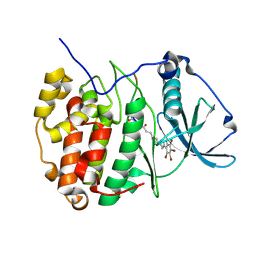 | | Structure of protein kinase CK2 catalytic subunit in complex with the CK2beta-competitive bisubstrate inhibitor ARC1502 | | Descriptor: | 8-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]octanoic acid, ARC1502, Casein kinase II subunit alpha | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
6SPW
 
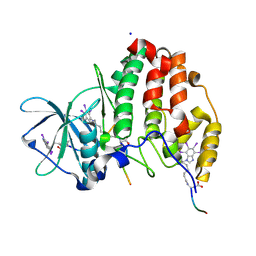 | | Structure of protein kinase CK2 catalytic subunit with the CK2beta-competitive bisubstrate inhibitor ARC3140 | | Descriptor: | 8-[4,5,6,7-tetrakis(iodanyl)benzimidazol-1-yl]octanoic acid, ARC3140, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
8IG0
 
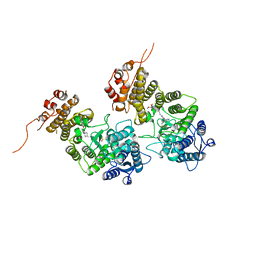 | | Crystal structure of menin in complex with DS-1594b | | Descriptor: | (1R,2S,4R)-4-[[4-(5,6-dimethoxypyridazin-3-yl)phenyl]methylamino]-2-[methyl-[6-[2,2,2-tris(fluoranyl)ethyl]thieno[2,3-d]pyrimidin-4-yl]amino]cyclopentan-1-ol, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Suzuki, M, Yoneyama, T, Imai, E. | | Deposit date: | 2023-02-20 | | Release date: | 2023-03-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel Menin-MLL1 inhibitor, DS-1594a, prevents the progression of acute leukemia with rearranged MLL1 or mutated NPM1.
Cancer Cell Int, 23, 2023
|
|
8QCD
 
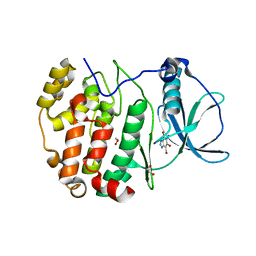 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR 4,5,6,7-TETRABROMOBENZOTRIAZOLE | | Descriptor: | 1,2-ETHANEDIOL, 4,5,6,7-TETRABROMOBENZOTRIAZOLE, Casein kinase II subunit alpha' | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
8QCG
 
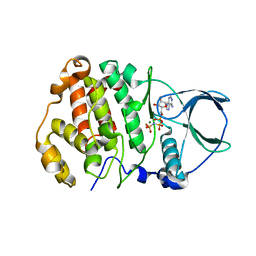 | |
8QBU
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR CX-4945 AND THE ALPHA-D-POCKET LIGAND 3,4-DICHLORO PHENETHYLAMINE (DPA) | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, ... | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
8QF1
 
 | |
8Q9S
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR SGC-CK2-1 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha', ~{N}-[5-[[3-cyano-7-(cyclopropylamino)-3~{H}-pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-methyl-phenyl]propanamide | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
7Z62
 
 | | Structure of the LecA lectin from Pseudomonas aeruginosa in complex with a biaryl-thiogalactoside | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-[4-[1-(4-methoxyphenyl)ethenyl]phenyl]sulfanyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Varrot, A. | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of potent 1,1-diarylthiogalactoside glycomimetic inhibitors of Pseudomonas aeruginosa LecA with antibiofilm properties.
Eur.J.Med.Chem., 247, 2022
|
|
2P3B
 
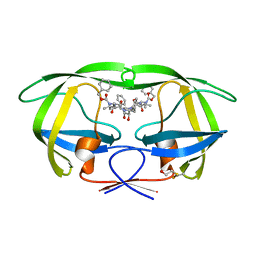 | | Crystal Structure of the subtype B wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
7Z63
 
 | |
2P3C
 
 | | Crystal Structure of the subtype F wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | ACETIC ACID, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
5XOD
 
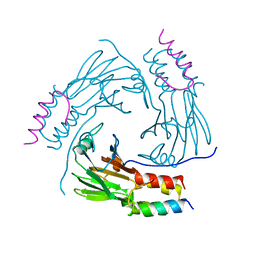 | | Crystal structure of human Smad2-Ski complex | | Descriptor: | Mothers against decapentaplegic homolog 2, Ski oncogene | | Authors: | Miyazono, K, Moriwaki, S, Ito, T, Tanokura, M. | | Deposit date: | 2017-05-27 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Sci Signal, 11, 2018
|
|
5IHL
 
 | |
6ENQ
 
 | | Structure of human PPAR gamma LBD in complex with Lanifibranor (IVA337) | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Boubia, B, Poupardin, O, Barth, M, Amaudrut, J, Broqua, P, Tallandier, M, Zeyer, D. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, Synthesis, and Evaluation of a Novel Series of Indole Sulfonamide Peroxisome Proliferator Activated Receptor (PPAR) alpha / gamma / delta Triple Activators: Discovery of Lanifibranor, a New Antifibrotic Clinical Candidate.
J. Med. Chem., 61, 2018
|
|
4UUY
 
 | | Structural Identification of the Vps18 beta-propeller reveals a critical role in the HOPS complex stability and function. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Behrmann, H, Gohlke, U, Heinemann, U. | | Deposit date: | 2014-08-01 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Identification of the Vps18 Beta-Propeller Reveals a Critical Role in the Hops Complex Stability and Function.
J.Biol.Chem., 289, 2014
|
|
