8PXP
 
 | |
1MQQ
 
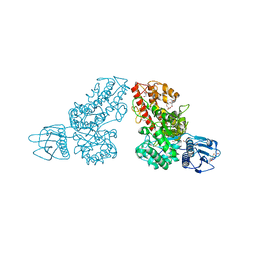 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL, alpha-D-glucopyranuronic acid | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1MQR
 
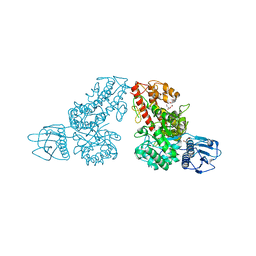 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE (E386Q) FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1MQP
 
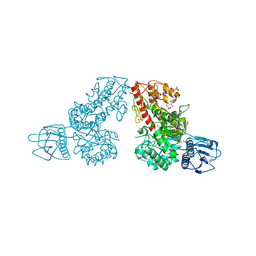 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1HIZ
 
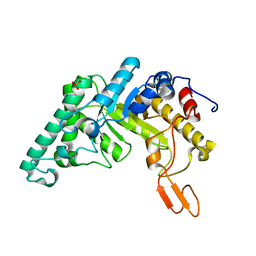 | | Xylanase T6 (Xt6) from Bacillus Stearothermophilus | | Descriptor: | ENDO-1,4-BETA-XYLANASE, SULFATE ION, alpha-D-galactopyranose, ... | | Authors: | Sainz, G, Tepplitsky, A, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-01-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Determination of the Extracellular Xylanase from Geobacillus Stearothermophilus by Selenomethionyl MAD Phasing
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZNW
 
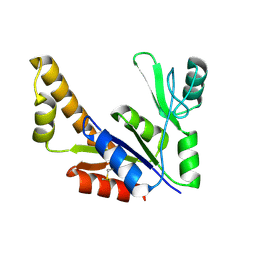 | | Crystal Structure Of Unliganded Form Of Mycobacterium tuberculosis Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Hible, G, Christova, P, Renault, L, Seclaman, E, Thompson, A, Girard, E, Munier-Lehmann, H, Cherfils, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase
Proteins, 62, 2006
|
|
5AOS
 
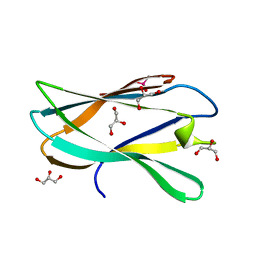 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1ZNZ
 
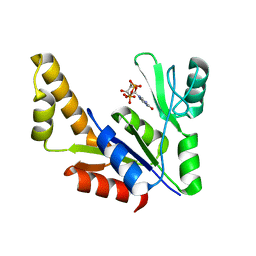 | | Crystal Structure Of The Reduced Form Of Mycobacterium tuberculosis Guanylate Kinase In Complex With GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanylate kinase | | Authors: | Hible, G, Christova, P, Renault, L, Seclaman, E, Thompson, A, Girard, E, Munier-Lehmann, H, Cherfils, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase
Proteins, 62, 2006
|
|
1ZNX
 
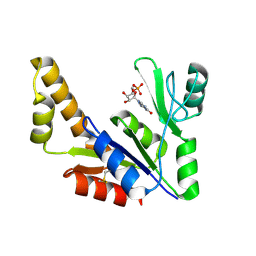 | | Crystal Structure Of Mycobacterium tuberculosis Guanylate Kinase In Complex With GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase | | Authors: | Hible, G, Christova, P, Renault, L, Seclaman, E, Thompson, A, Girard, E, Munier-Lehmann, H, Cherfils, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase
Proteins, 62, 2006
|
|
1ZNY
 
 | | Crystal Structure Of Mycobacterium tuberculosis Guanylate Kinase In Complex With GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanylate kinase | | Authors: | Hible, G, Christova, P, Renault, L, Seclaman, E, Thompson, A, Girard, E, Munier-Lehmann, H, Cherfils, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase
Proteins, 62, 2006
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1QUN
 
 | | X-RAY STRUCTURE OF THE FIMC-FIMH CHAPERONE ADHESIN COMPLEX FROM UROPATHOGENIC E.COLI | | Descriptor: | MANNOSE-SPECIFIC ADHESIN FIMH, PAPD-LIKE CHAPERONE FIMC | | Authors: | Choudhury, D, Thompson, A, Stojanoff, V, Langerman, S, Pinkner, J, Hultgren, S.J, Knight, S. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Science, 285, 1999
|
|
5EPV
 
 | | Histidine kinase domain from the LOV-HK blue-light receptor from Brucella abortus | | Descriptor: | Blue-light-activated histidine kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Rinaldi, J, Guimaraes, B.G, Legrand, P, Thompson, A, Paris, G, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the HWE Histidine Kinase Family: The Brucella Blue Light-Activated Histidine Kinase Domain.
J.Mol.Biol., 428, 2016
|
|
1JU2
 
 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
1K9F
 
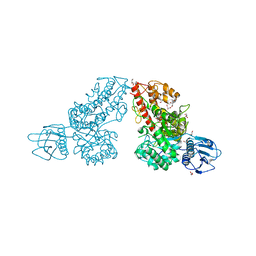 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K9E
 
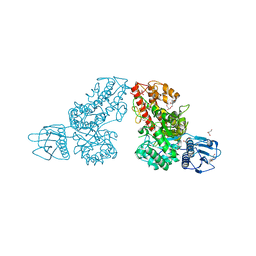 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with 4-O-methyl-glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K9D
 
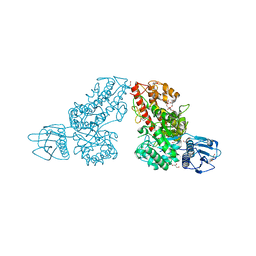 | | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1NCG
 
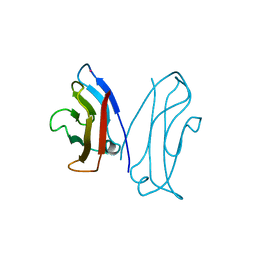 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCI
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
4AII
 
 | | Crystal structure of the rat REM2 GTPase - G domain bound to GDP | | Descriptor: | GTP-BINDING PROTEIN REM 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Reymond, P, Coquard, A, Chenon, M, Zeghouf, M, El Marjou, A, Thompson, A, Menetrey, J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Gdp-Bound G Domain of the Rgk Protein Rem2.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1GQU
 
 | |
1JOT
 
 | | STRUCTURE OF THE LECTIN MPA COMPLEXED WITH T-ANTIGEN DISACCHARIDE | | Descriptor: | AGGLUTININ, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Lee, X, Thompson, A, Zhang, Z, Hoa, T.-T, Biesterfeldt, J, Ogata, C, Xu, L, Johnston, R.A.Z, Young, N.M. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc.
J.Biol.Chem., 273, 1998
|
|
4CA4
 
 | | Crystal structure of FimH lectin domain with the Tyr48Ala mutation, in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FIMH, heptyl alpha-D-mannopyranoside | | Authors: | Rabbani, S, Bouckaert, J, Zalewski, A, Preston, R, Eid, S, Thompson, A, Puorger, C, Glockshuber, R, Ernst, B. | | Deposit date: | 2013-10-06 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Mutation of Tyr137 of the universal Escherichia coli fimbrial adhesin FimH relaxes the tyrosine gate prior to mannose binding.
IUCrJ, 4, 2017
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
