2K58
 
 | |
2JST
 
 | | Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets II: Halothane Effects on Structure and Dynamics | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, Four-Alpha-Helix Bundle | | Authors: | Cui, T, Bondarenko, V, Ma, D, Canlas, C, Brandon, N.R, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2007-07-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part II: halothane effects on structure and dynamics
Biophys.J., 94, 2008
|
|
2K1E
 
 | | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA | | Descriptor: | water soluble analogue of potassium channel, KcsA | | Authors: | Ma, D, Xu, Y, Tillman, T, Tang, P, Meirovitch, E, Eckenhoff, R, Carnini, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2MAW
 
 | |
2KB1
 
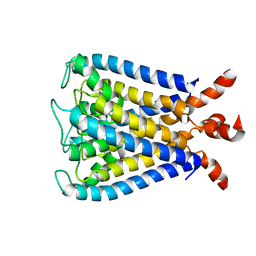 | | NMR studies of a channel protein without membrane: structure and dynamics of water-solubilized KcsA | | Descriptor: | WSK3 | | Authors: | Ma, D, Xu, Y, Tillman, T, Tang, P, Meirovitch, E, Eckenhoff, R, Carnini, A. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2K59
 
 | |
2LLY
 
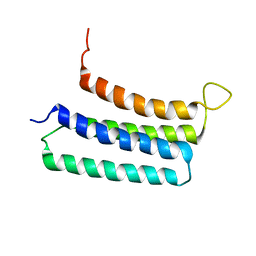 | | NMR structures of the transmembrane domains of the nAChR a4 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit alpha-4 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
2LKH
 
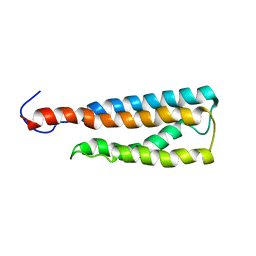 | | WSA minor conformation | | Descriptor: | Acetylcholine receptor | | Authors: | Xu, Y, Mowrey, D, Cui, T, Perez-Aguilar, J, Saven, J.G, Eckenhoff, R, Tang, P. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor.
Biochim.Biophys.Acta, 1818, 2011
|
|
2LM2
 
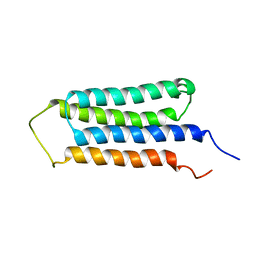 | | NMR structures of the transmembrane domains of the AChR b2 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit beta-2 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
2LKG
 
 | | WSA major conformation | | Descriptor: | Acetylcholine receptor | | Authors: | Xu, Y, Mowrey, D, Cui, T, Perez-Aguilar, J.M, Saven, J.G, Eckenhoff, R, Tang, P. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor.
Biochim.Biophys.Acta, 1818, 2011
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
3ME2
 
 | | Crystal structure of mouse RANKL-RANK complex | | Descriptor: | CHLORIDE ION, SODIUM ION, Tumor necrosis factor ligand superfamily member 11, ... | | Authors: | Walter, S.W, Liu, C.Z, Zhu, X.K, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
3ME4
 
 | | Crystal structure of mouse RANK | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walter, S.W, Liu, C, Zhu, X, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
8Z98
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z90
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8N
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z97
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8J
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9R
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
