3U6Y
 
 | |
2E85
 
 | | Crystal Structure of the Hydrogenase 3 Maturation protease | | Descriptor: | CALCIUM ION, Hydrogenase 3 maturation protease | | Authors: | Tanaka, T, Kumarevel, T.S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of hydrogenase maturating endopeptidase HycI from Escherichia coli
Biochem.Biophys.Res.Commun., 389, 2009
|
|
2H09
 
 | |
4W4Q
 
 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
8ZLR
 
 | | SFX structure of CraCRY in the semiquinone state | | Descriptor: | Cryptochrome photoreceptor, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-05-21 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
2IE1
 
 | | Polyamines stabilize left-handed Z-DNA. We found new type of polyamine which stabilize left-handed Z-DNA by X-ray crystallography | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), N-(2-AMINOETHYL)-N'-{2-[(2-AMINOETHYL)AMINO]ETHYL}ETHANE-1,2-DIAMINE | | Authors: | Ohishi, H, Odoko, M, Tsukamoto, K, Hiyama, Y, Maezaki, N, Grzeskowiak, K, Ishida, T, Tanaka, T, Okabe, N, Fukuyama, K. | | Deposit date: | 2006-09-16 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Polyamines stabilize left-handed Z-DNA. We found new type of polyamine which stabilize left-handed Z-DNA by X-ray crystallography
To be Published
|
|
1TO4
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | COPPER (II) ION, Superoxide dismutase, ZINC ION | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TO5
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase, ... | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
9J7W
 
 | | Channel Rhodospin from Klebsormidium nitens (KnChR) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, KnChR, RETINAL | | Authors: | Wang, Y.Z, Akasaka, H, Tanaka, T, Sano, F.K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-08-20 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure of a blue-shifted channelrhodopsin from Klebsormidium nitens.
Nat Commun, 16, 2025
|
|
413D
 
 | | A'-FORM RNA DOUBLE HELIX IN THE SINGLE CRYSTAL STRUCTURE OF R(UGAGCUUCGGCUC) | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*UP*UP*CP*GP*GP*CP*UP*C)-3') | | Authors: | Tanaka, Y, Fujii, S, Hiroaki, H, Sakata, T, Tanaka, T, Uesugi, S, Tomita, K.-I, Kyogoku, Y. | | Deposit date: | 1998-07-10 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A'-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC).
Nucleic Acids Res., 27, 1999
|
|
5H2L
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 5.25 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2N
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 95.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2O
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3WXQ
 
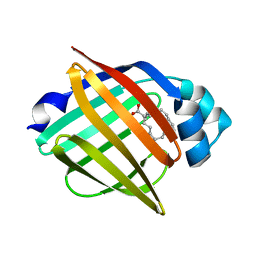 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WXS
 
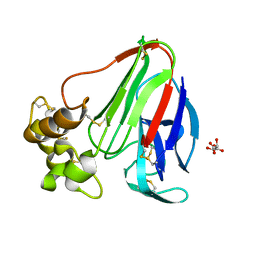 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
9LRE
 
 | | Cryo-EM structure of the histamine H4 receptor-Gi protein complex (Overall) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptor
To Be Published
|
|
9LRB
 
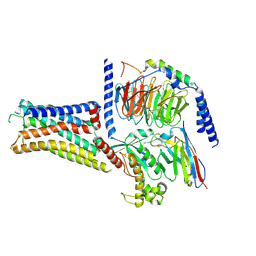 | | Cryo-EM structure of the histamine H1 receptor-Gs protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LRD
 
 | | Cryo-EM structure of the histamine H1 receptor-Gi protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LRC
 
 | | Cryo-EM structure of the histamine H4 receptor-Gi protein complex (Receptor focused) | | Descriptor: | HISTAMINE, Histamine H4 receptor,Genome polyprotein | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
6G7L
 
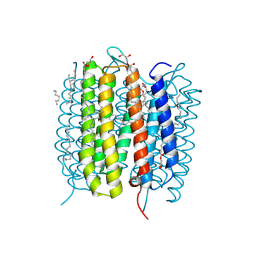 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 8.3 ms state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7H
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: resting state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7J
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 457-646 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
