4H0O
 
 | |
5AX2
 
 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
6KFQ
 
 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
8IIA
 
 | |
8IGU
 
 | |
8IGW
 
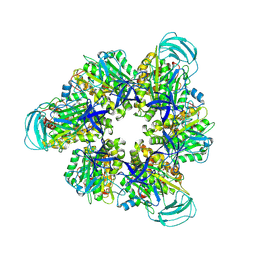 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
7COQ
 
 | |
3VY9
 
 | | Crystal structure of PorB from Neisseria meningitidis in complex with cesium ion, space group H32 | | Descriptor: | CESIUM ION, Outer membrane protein | | Authors: | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3VY8
 
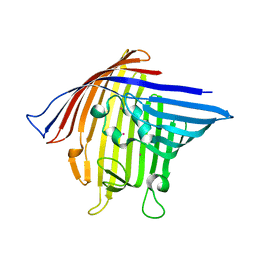 | | Crystal Structure of PorB from Neisseria meningitidis in complex with Cesium ion, space group P63 | | Descriptor: | CESIUM ION, Outer membrane protein | | Authors: | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3WI4
 
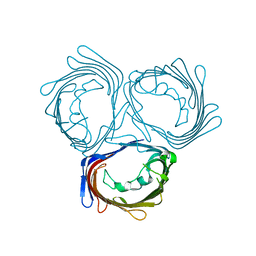 | | Crystal structure of wild-type PorB from Neisseria meningitidis serogroup B | | Descriptor: | Major outer membrane protein P.IB | | Authors: | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
3WI5
 
 | | Crystal structure of the Loop 7 mutant PorB from Neisseria meningitidis serogroup B | | Descriptor: | CITRATE ANION, Major outer membrane protein P.IB | | Authors: | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
1IT8
 
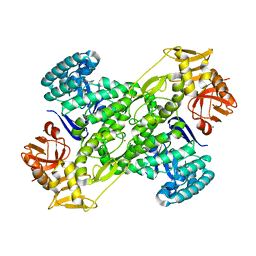 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IQ8
 
 | | Crystal Structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii | | Descriptor: | ARCHAEOSINE TRNA-GUANINE TRANSGLYCOSYLASE, MAGNESIUM ION, ZINC ION | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
6JNO
 
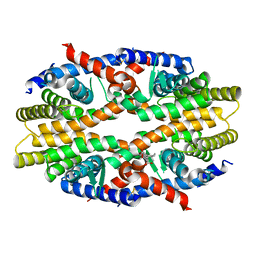 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
4D7Y
 
 | | Crystal structure of mouse C1QL1 globular domain | | Descriptor: | C1Q-RELATED FACTOR, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kakegawa, W, Mitakidis, N, Miura, E, Abe, M, Matsuda, K, Takeo, Y, Kohda, K, Motohashi, J, Takahashi, A, Nagao, S, Muramatsu, S, Watanabe, M, Sakimura, K, Aricescu, A.R, Yuzaki, M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Anterograde C1Ql1 Signaling is Required in Order to Determine and Maintain a Single-Winner Climbing Fiber in the Mouse Cerebellum
Neuron, 85, 2015
|
|
5ZN7
 
 | |
6JOW
 
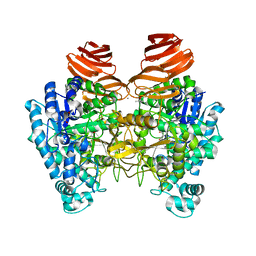 | |
6KFG
 
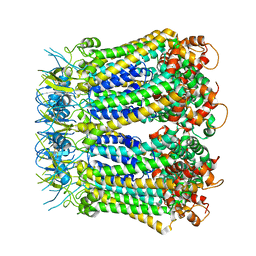 | | Undocked INX-6 hemichannel in detergent | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
2KWC
 
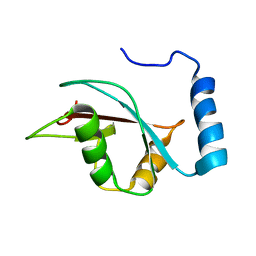 | | The NMR structure of the autophagy-related protein Atg8 | | Descriptor: | Autophagy-related protein 8 | | Authors: | Kumeta, H, Watanabe, M, Nakatogawa, H, Yamaguchi, M, Ogura, K, Adachi, W, Fujioka, Y, Noda, N.N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the autophagy-related protein Atg8
J.Biomol.Nmr, 47, 2010
|
|
8HX6
 
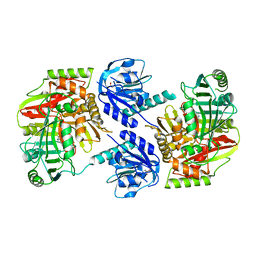 | |
8HX7
 
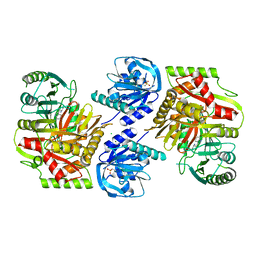 | |
8HX9
 
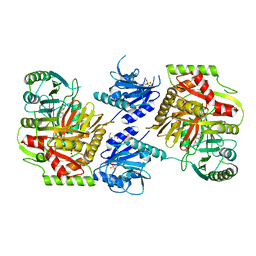 | | Crystal structure of 4-amino-4-deoxychorismate synthase from Streptomyces venezuelae with chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-amino-4-deoxychorismate synthase, FORMIC ACID, ... | | Authors: | Nakamichi, Y, Watanabe, M. | | Deposit date: | 2023-01-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the allosteric pathway of 4-amino-4-deoxychorismate synthase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8HX8
 
 | |
