5H83
 
 | | HETEROYOHIMBINE SYNTHASE HYS FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase HYS | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
5H82
 
 | | HETEROYOHIMBINE SYNTHASE THAS2 FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase THAS2 | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
4XHC
 
 | | rhamnosidase from Klebsiella oxytoca with rhamnose bound | | Descriptor: | Alpha-L-rhamnosidase, SULFATE ION, alpha-L-rhamnopyranose | | Authors: | O'Neill, E.O, Stevenson, C.E.M, Patterson, M.J, Rejzek, M, Chauvin, A, Lawson, D.M, Field, R.A. | | Deposit date: | 2015-01-05 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a novel two domain GH78 family alpha-rhamnosidase from Klebsiella oxytoca with rhamnose bound.
Proteins, 83, 2015
|
|
2WMC
 
 | | Crystal structure of eukaryotic initiation factor 4E from Pisum sativum | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Ashby, J.A, Stevenson, C.E.M, Maule, A.J, Lawson, D.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Mutational Analysis of Eif4E in Relation to Sbm1 Resistance to Pea Seed-Borne Mosaic Virus in Pea.
Plos One, 6, 2011
|
|
4CKK
 
 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
4CKL
 
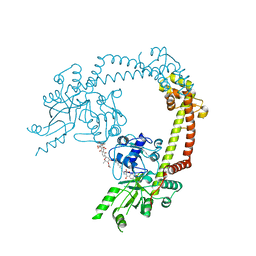 | | Structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit with simocyclinone D8 bound | | Descriptor: | DNA GYRASE SUBUNIT A, SIMOCYCLINONE D8 | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
5L40
 
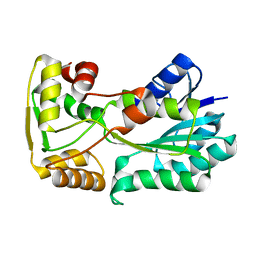 | | polyketide ketoreductase SimC7 - apo crystal form 1 | | Descriptor: | polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5L4L
 
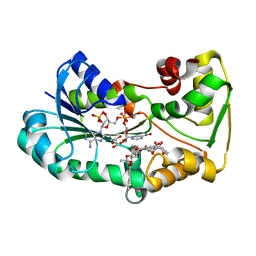 | | polyketide ketoreductase SimC7 - ternary complex with NADP+ and 7-oxo-SD8 | | Descriptor: | 7-oxo-simocyclinone D8, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5L3Z
 
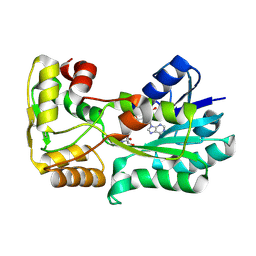 | | polyketide ketoreductase SimC7 - binary complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5L45
 
 | | polyketide ketoreductase SimC7 - apo crystal form 2 | | Descriptor: | GLYCEROL, SimC7, TETRAETHYLENE GLYCOL | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5LGV
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5LQD
 
 | | Trehalose-6-phosphate synthase, GDP-glucose-dependent OtsA | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase | | Authors: | Miah, F, Asencion Diez, M.D, Stevenson, C.E.M, Lawson, D.M, Iglesias, A.A, Bornemann, S. | | Deposit date: | 2016-08-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Production and Utilization of GDP-glucose in the Biosynthesis of Trehalose 6-Phosphate by Streptomyces venezuelae.
J. Biol. Chem., 292, 2017
|
|
5LGW
 
 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant co-crystallised with maltodextrin | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5M0T
 
 | | Alpha-ketoglutarate-dependent non-heme iron oxygenase EasH | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent non-heme iron oxygenase EasH, FE (II) ION, ... | | Authors: | Jakubczyk, D, Caputi, L, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2016-10-05 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of EasH (Aspergillus japonicus) - an oxidase involved in cycloclavine biosynthesis.
Chem. Commun. (Camb.), 52, 2016
|
|
5A6W
 
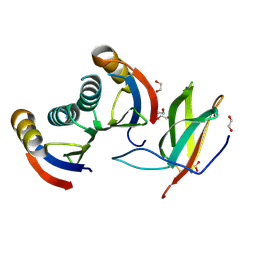 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5A6P
 
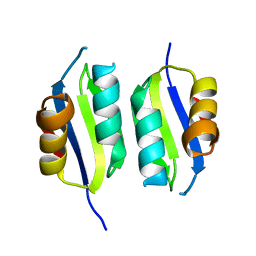 | | Heavy metal associated domain of NLR-type immune receptor Pikp1 from rice (Oryza sativa) | | Descriptor: | RESISTANCE PROTEIN PIKP-1 | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5CPS
 
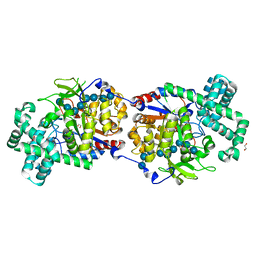 | | Disproportionating enzyme 1 from Arabidopsis - maltotriose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CPQ
 
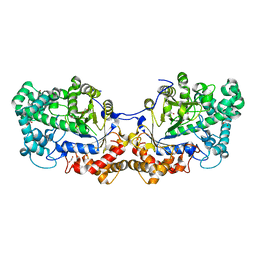 | | Disproportionating enzyme 1 from Arabidopsis - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
2XGI
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3,4- epoxybutyl alpha-D-glucopyranoside | | Descriptor: | (3R)-3-hydroxybutyl alpha-D-glucopyranoside, (3S)-3-hydroxybutyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, ... | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chemical genetics and cereal starch metabolism: structural basis of the non-covalent and covalent inhibition of barley beta-amylase.
Mol Biosyst, 7, 2011
|
|
2XGB
 
 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
8A3N
 
 | | Geissoschizine synthase from Catharanthus roseus - binary complex with NADP+ | | Descriptor: | Geissoschizine synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Langley, C, Tatsis, E, Hong, B, Nakamura, Y, Kamileen, M.O, Paetz, C, Stevenson, C.E.M, Basquin, J, Lawson, D.M, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8PFD
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFC
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
