3KE9
 
 | | Crystal structure of IspH:Intermediate-complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEL
 
 | | Crystal Structure of IspH:PP complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PYROPHOSPHATE 2- | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE8
 
 | | Crystal structure of IspH:HMBPP-complex | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8BJ7
 
 | |
8BJ8
 
 | |
6SG2
 
 | | FeFe Hydrogenase from Desulfovibrio desulfuricans in Hinact state | | Descriptor: | HydB, IRON/SULFUR CLUSTER, Periplasmic [Fe] hydrogenase large subunit, ... | | Authors: | Galle, L.M, Span, I. | | Deposit date: | 2019-08-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Caught in the H inact : Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O 2 -stable State of [FeFe] Hydrogenase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4EB3
 
 | | Crystal structure of IspH in complex with iso-HMBPP | | Descriptor: | 3-(hydroxymethyl)but-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Wang, W, Wang, K, Span, I, Bacher, A, Groll, M, Oldfield, E. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Are free radicals involved in IspH catalysis? An EPR and crystallographic investigation.
J.Am.Chem.Soc., 134, 2012
|
|
6YO5
 
 | | Crystal structure of the M295F variant of Ssl1 | | Descriptor: | ALA-HIS-ALA, COPPER (II) ION, Copper oxidase, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of the axial ligand mutation on spectral and structural properties of Ssl1 laccase
To Be Published
|
|
6Y4A
 
 | |
6YZF
 
 | | Crystal structure of the M295Y variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase, GLU-HIS-SER, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-05-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Effect of the axial ligand mutation on spectral and structural properties of Ssl1 laccase
To Be Published
|
|
6YZD
 
 | |
6YZY
 
 | |
6SQV
 
 | | Structure of the U1A variant A1-98 Y31H/Q36R/R70W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
6SQN
 
 | |
6SQT
 
 | |
6SR7
 
 | | Structure of the U1A variant A1-98 Y31H/Q36R/K98W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
6SQQ
 
 | |
7PDU
 
 | |
3F7T
 
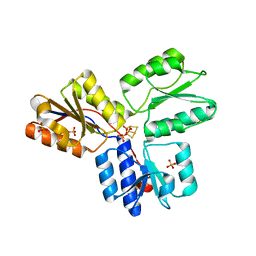 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3RJ6
 
 | |
