2IN3
 
 | | Crystal structure of a putative protein disulfide isomerase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein, ... | | Authors: | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-05 | | Release date: | 2006-11-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a putative protein disulfide isomerase from Nitrosomonas europaea
To be Published
|
|
2IGT
 
 | | Crystal Structure of the SAM Dependent Methyltransferase from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-25 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the SAM Dependent Methyltransferase from Agrobacterium tumefaciens
To be Published
|
|
1Y0U
 
 | | Crystal Structure of the putative arsenical resistance operon repressor from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, arsenical resistance operon repressor, putative | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-28 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the putative arsenical resistance operon repressor from Archaeoglobus fulgidus
To be Published
|
|
2IJL
 
 | | The structure of a putative ModE from Agrobacterium tumefaciens. | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum-binding transcriptional repressor, SULFATE ION | | Authors: | Cuff, M.E, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative ModE from Agrobacterium tumefaciens.
To be Published
|
|
3NKE
 
 | | High resolution structure of the C-terminal domain CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SULFITE ION, ... | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
2IGS
 
 | | Crystal Structure of the Protein of Unknown Function from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, GLYCEROL, Hypothetical protein, ... | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-25 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Pseudomonas aeruginosa
To be Published
|
|
1Y0N
 
 | | Structure of Protein of Unknown Function PA3463 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Hypothetical UPF0270 protein PA3463 | | Authors: | Binkowski, T.A, Edwards, A, Savchenko, A, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein PA3463 from Pseudomonas aeruginosa strain PAO1
To be Published
|
|
1Y88
 
 | | Crystal Structure of Protein of Unknown Function AF1548 | | Descriptor: | CHLORIDE ION, Hypothetical protein AF1548, SULFATE ION | | Authors: | Lunin, V.V, Evdokimova, E, Kudritskaya, M, Cuff, M.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of hypothetical protein AF1548 from Archaeoglobus fulgidus
To be Published
|
|
1Y0K
 
 | | Structure of Protein of Unknown Function PA4535 from Pseudomonas aeruginosa strain PAO1, Monooxygenase Superfamily | | Descriptor: | hypothetical protein PA4535 | | Authors: | Nocek, B.P, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-18 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 A Crystal Structure of the Hypothetical Protein Pa4535 from Pseudomonas Aeruginosa
To be Published
|
|
1Y7P
 
 | | 1.9 A Crystal Structure of a Protein of Unknown Function AF1403 from Archaeoglobus fulgidus, Probable Metabolic Regulator | | Descriptor: | Hypothetical protein AF1403, ZINC ION, beta-D-ribopyranose | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of a hypothetical protein AF1403 from Archaeoglobus fulgidus
To be Published
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
7SNU
 
 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | Descriptor: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
3ERP
 
 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-02 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
3EXM
 
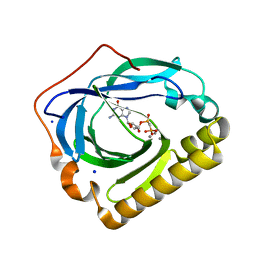 | | Crystal structure of the phosphatase SC4828 with the non-hydrolyzable nucleotide GPCP | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID GUANOSYL ESTER, ... | | Authors: | Singer, A.U, Xu, X, Zheng, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of a new family of prokaryotic nucleoside diphosphatases.
To be Published
|
|
3EXC
 
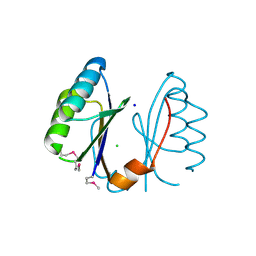 | | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Singer, A.U, Skarina, T, Tan, K, Kagan, O, Onopriyenko, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus
To be Published
|
|
3FIW
 
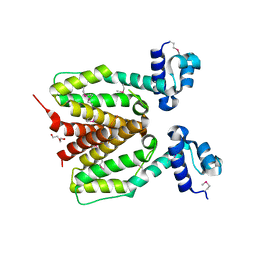 | | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative tetR-family transcriptional regulator | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
3NA2
 
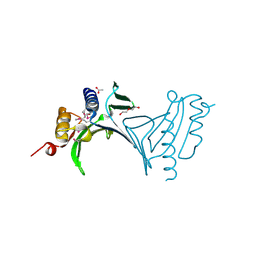 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
2IKS
 
 | | Crystal structure of N-terminal truncated DNA-binding transcriptional dual regulator from Escherichia coli K12 | | Descriptor: | DNA-binding transcriptional dual regulator | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of N-terminal truncated DNA-binding transcriptional
dual regulator from Escherichia coli K12
To be Published
|
|
6CZP
 
 | | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN.
To Be Published
|
|
4E0B
 
 | | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6 | | Descriptor: | ACETATE ION, Malate dehydrogenase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-02 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6
To be Published
|
|
7TBU
 
 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
6D33
 
 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
4WRP
 
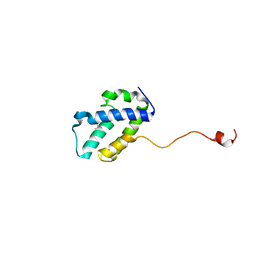 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
