4IQL
 
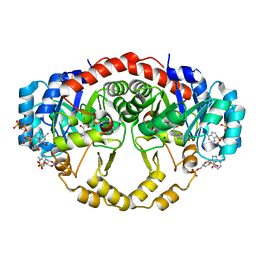 | | Crystal Structure of Porphyromonas gingivalis Enoyl-ACP Reductase II (FabK) with cofactors NADPH and FMN | | Descriptor: | Enoyl-(Acyl-carrier-protein) reductase II, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Hevener, K.E, Santarsiero, B.D, Su, P.-C, Boci, T, Truong, K, Johnson, M.E, Mehboob, S. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural characterization of Porphyromonas gingivalis enoyl-ACP reductase II (FabK).
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4J4T
 
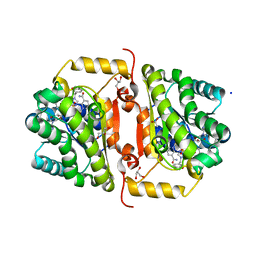 | | Crystal Structure of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold | | Descriptor: | 1-(1,3-benzodioxol-5-ylmethyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4J3F
 
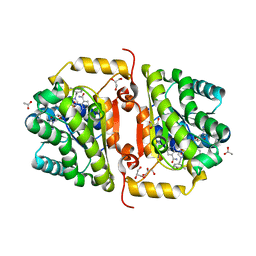 | | Crystal Structure of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold. | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2QX6
 
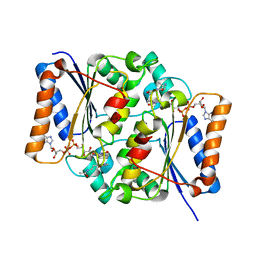 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QWX
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QTN
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Sershon, V.C, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and X-ray structural evidence for negative cooperativity in substrate binding to nicotinate mononucleotide adenylyltransferase (NMAT) from Bacillus anthracis.
J.Mol.Biol., 385, 2009
|
|
2QX8
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QTM
 
 | |
2QX9
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QTR
 
 | |
2QIO
 
 | |
2QX4
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
4NZ9
 
 | | Crystal Structure of FabI from S. aureus in complex with a novel benzimidazole inhibitor | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mehboob, S, Johnson, M.E, Santarsiero, B.D. | | Deposit date: | 2013-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To be Published
|
|
4PT5
 
 | |
3F4C
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10, with glycerol bound | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3F4D
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10 | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
5IRO
 
 | |
6CN8
 
 | | High-resolution structure of ClpC1-NTD binding to Rufomycin-I | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC1, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Abad-Zapatero, C, Wolf, N.W. | | Deposit date: | 2018-03-07 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structure of ClpC1-Rufomycin and Ligand Binding Studies Provide a Framework to Design and Optimize Anti-Tuberculosis Leads.
Acs Infect Dis., 5, 2019
|
|
4Q18
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 4 [1-[5-(4-{[(2,6-diaminopyrimidin-4-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol] | | Descriptor: | 1-(5-(4-(((2,6-diaminopyrimidin-4-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenoxy)-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
1AXS
 
 | | MATURE OXY-COPE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | (1S,2S,5S)2-(4-GLUTARIDYLBENZYL)-5-PHENYL-1-CYCLOHEXANOL, CADMIUM ION, OXY-COPE CATALYTIC ANTIBODY | | Authors: | Mundorff, E.C, Ulrich, H.D, Stevens, R.C. | | Deposit date: | 1997-10-20 | | Release date: | 1998-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interplay between binding energy and catalysis in the evolution of a catalytic antibody.
Nature, 389, 1997
|
|
1PHZ
 
 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
2PHM
 
 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
7TMK
 
 | |
7TLU
 
 | |
7TMH
 
 | |
