7CIE
 
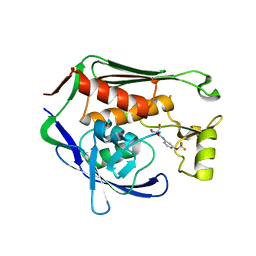 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-3-oxidanyl-N-[3-(trifluoromethyloxy)phenyl]propanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIA
 
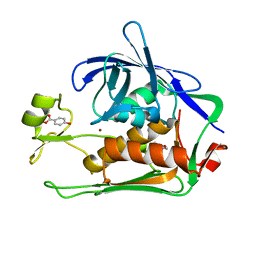 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7WBT
 
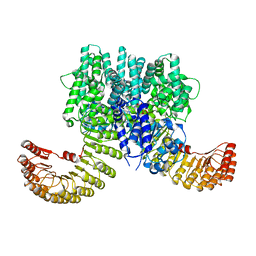 | | Crystal structure of bovine NLRP9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Kamitsukasa, Y, Shimizu, T, Ohto, U. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of NLRP9 reveals a unique C-terminal region with putative regulatory function.
Febs Lett., 596, 2022
|
|
7WBU
 
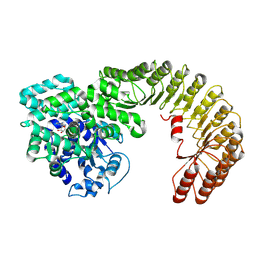 | | Cryo-EM structure of bovine NLRP9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Kamitsukasa, Y, Shimizu, T, Ohto, U. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The structure of NLRP9 reveals a unique C-terminal region with putative regulatory function.
Febs Lett., 596, 2022
|
|
2DVS
 
 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVR
 
 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVQ
 
 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2E3K
 
 | |
2DVW
 
 | |
2DWZ
 
 | |
