2NYN
 
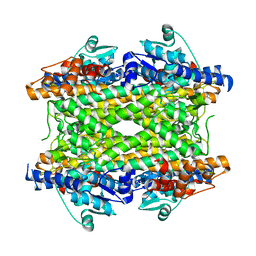 | | Crystal structure of phenylalanine ammonia-lyase from Anabaena variabilis | | Descriptor: | Phenylalanine/histidine ammonia-lyase | | Authors: | Louie, G.V, Moffitt, M.C, Bowman, M.E, Pence, J, Noel, J.P, Moore, B.S. | | Deposit date: | 2006-11-21 | | Release date: | 2007-02-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Two Cyanobacterial Phenylalanine Ammonia Lyases: Kinetic and Structural Characterization.
Biochemistry, 46, 2007
|
|
2O7D
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides, complexed with caffeate | | Descriptor: | CAFFEIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
2O7F
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides (His89Phe variant), complexed with coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-11 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
1U0M
 
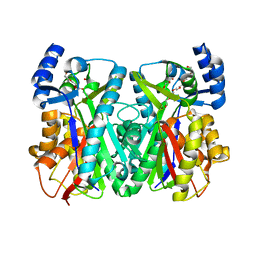 | | Crystal Structure of 1,3,6,8-Tetrahydroxynaphthalene Synthase (THNS) from Streptomyces coelicolor A3(2): a Bacterial Type III Polyketide Synthase (PKS) Provides Insights into Enzymatic Control of Reactive Polyketide Intermediates | | Descriptor: | GLYCEROL, POLYETHYLENE GLYCOL (N=34), putative polyketide synthase | | Authors: | Austin, M.B, Izumikawa, M, Bowman, M.E, Udwary, D.W, Ferrer, J.L, Moore, B.S, Noel, J.P. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates
J.Biol.Chem., 279, 2004
|
|
4XLO
 
 | | Crystal Structure of EncM (crystallized with 4 mM NADPH) | | Descriptor: | FAD-dependent oxygenase EncM, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Teufel, R. | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
8TWN
 
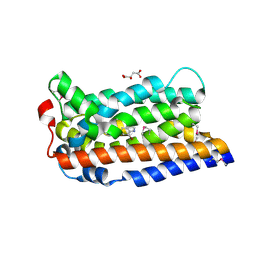 | |
8TWT
 
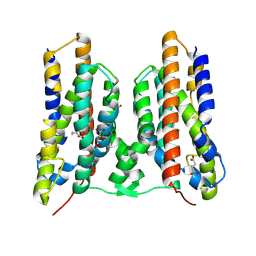 | |
8TWW
 
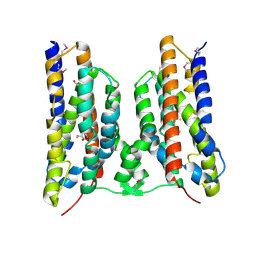 | |
6CY8
 
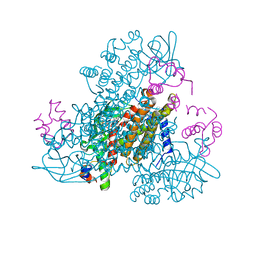 | | Crystal structure of FAD-dependent dehydrogenase | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Alpha/beta hydrolase fold protein, Butyryl-CoA dehydrogenase, ... | | Authors: | Agarwal, V. | | Deposit date: | 2018-04-05 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Biochemistry, 58, 2019
|
|
6CXT
 
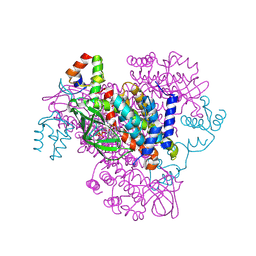 | | Crystal structure of FAD-dependent dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase fold protein, Butyryl-CoA dehydrogenase, ... | | Authors: | Agarwal, V. | | Deposit date: | 2018-04-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Biochemistry, 58, 2019
|
|
2WR8
 
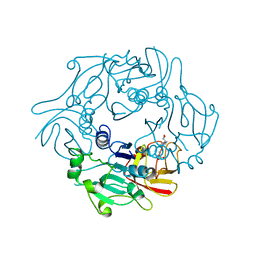 | | Structure of Pyrococcus horikoshii SAM hydroxide adenosyltransferase in complex with SAH | | Descriptor: | PUTATIVE UNCHARACTERIZED PROTEIN PH0463, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMahon, S.A, Deng, H, O'Hagan, D, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Mechanistic Insights Into Water Activation in Sam Hydroxide Adenosyltransferase (Duf-62).
Chembiochem, 10, 2009
|
|
4HNP
 
 | | Crystal structure of yeast 20S proteasome in complex with vinylketone carmaphycin analogue VNK1 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylhept-1-en-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-20 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4HRD
 
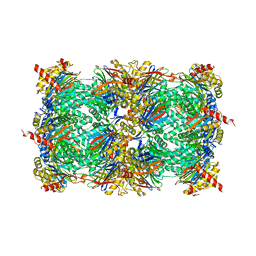 | | Crystal structure of yeast 20S proteasome in complex with the natural product carmaphycin A | | Descriptor: | N-[(2S)-1-({(2S)-1-{[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]amino}-4-[(R)-methylsulfinyl]-1-oxobutan-2-yl}amino)-3-methyl-1-oxobutan-2-yl]hexanamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-27 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4HRC
 
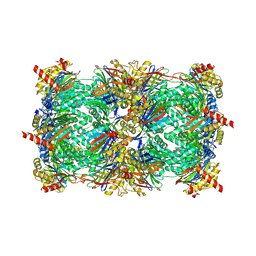 | | Crystal structure of yeast 20S proteasome in complex with epoxyketone carmaphycin analogue 3 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-27 | | Release date: | 2014-01-29 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
4LTC
 
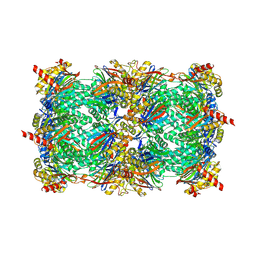 | | Crystal structure of yeast 20S proteasome in complex with enone carmaphycin analogue 6 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(4S,5S,6R)-5-hydroxy-2,6-dimethyloctan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Stein, M, Trivella, D.B.B, Groll, M. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
6FY8
 
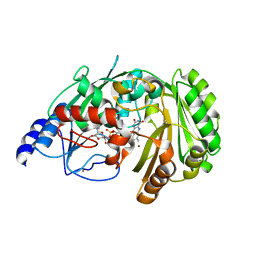 | | The crystal structure of EncM bromide soak | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYA
 
 | |
6FOQ
 
 | |
6FYE
 
 | | The crystal structure of EncM H138T mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYF
 
 | | The crystal structure of EncM V135M mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FP3
 
 | |
6FY9
 
 | |
6FYD
 
 | | The crystal structure of EncM T139V mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYG
 
 | | The crystal structure of EncM V135T mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7JPJ
 
 | | Crystal Structure of the essential dimeric LYSA from Phaeodactylum tricornutum | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, SULFATE ION | | Authors: | Fedorov, E, Belinski, V.A, Brunson, J.K, Almo, S.C, Dupont, C.L, Ghosh, A. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Phaeodactylum tricornutum diaminopimelate decarboxylase was acquired via horizontal gene transfer from bacteria and displays substrate promiscuity
Biorxiv, 2020
|
|
