4ZM7
 
 | |
7YK9
 
 | | Neutron Structure of PcyA I86D Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Unno, M, Igarashi, K. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
7YKB
 
 | | Neutron Structure of PcyA D105N Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase, SODIUM ION | | Authors: | Unno, M, Nanasawa, R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.38 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
7YL8
 
 | |
7WNP
 
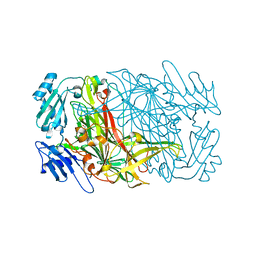 | |
7WNO
 
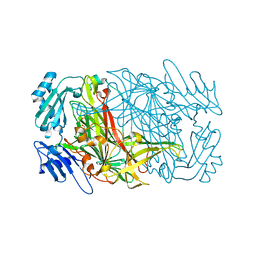 | |
7XVX
 
 | |
8W6X
 
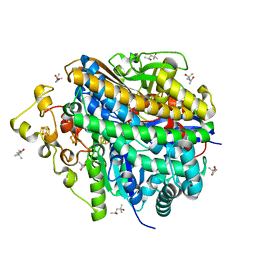 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8J6G
 
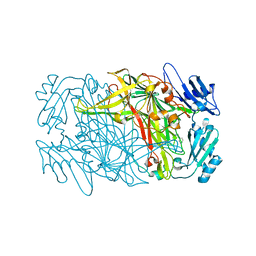 | |
8Z57
 
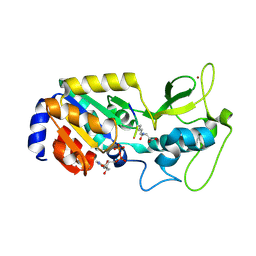 | |
8Z55
 
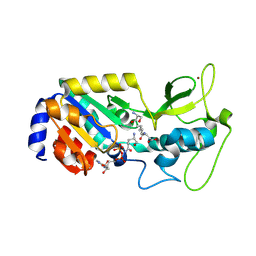 | |
8Z58
 
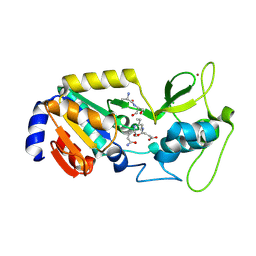 | |
8Z54
 
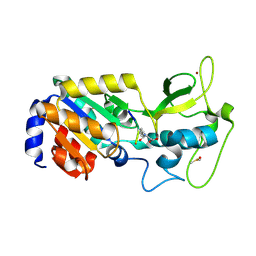 | |
8Z56
 
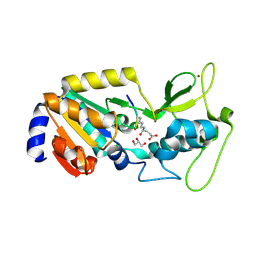 | |
8IE8
 
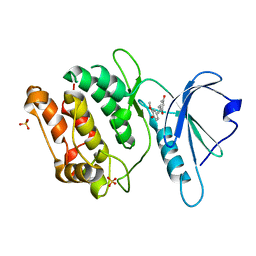 | | Crystal structure of DAPK1 in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE7
 
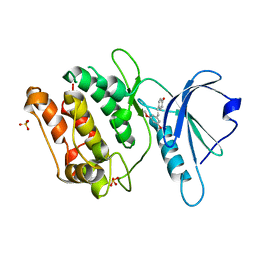 | | Crystal structure of DAPK1 in complex with pterostilbene | | Descriptor: | Death-associated protein kinase 1, Pterostilbene, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE6
 
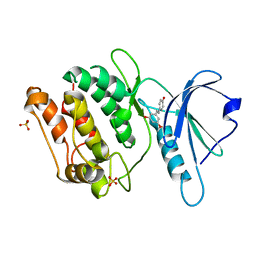 | | Crystal structure of DAPK1 in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE5
 
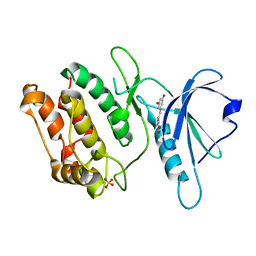 | | Crystal structure of DAPK1 in complex with oxyresveratrol | | Descriptor: | Death-associated protein kinase 1, SULFATE ION, trans-oxyresveratrol | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IG1
 
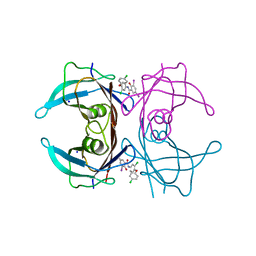 | | Crystal structure of wild-type transthyretin in complex with rafoxanide | | Descriptor: | SODIUM ION, Transthyretin, ~{N}-[3-chloranyl-4-(4-chloranylphenoxy)phenyl]-3,5-bis(iodanyl)-2-oxidanyl-benzamide | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Rafoxanide, a salicylanilide anthelmintic, interacts with human plasma protein transthyretin.
Febs J., 290, 2023
|
|
8W42
 
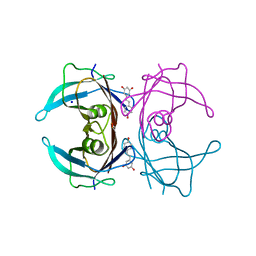 | | X-ray crystal structure of V30M-TTR in complex with resveratrol | | Descriptor: | RESVERATROL, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W44
 
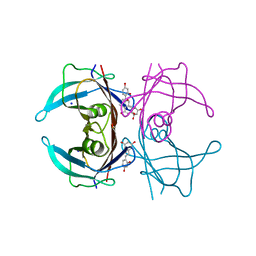 | | X-ray crystal structure of V30M-TTR in complex with oxyresveratrol | | Descriptor: | SODIUM ION, Transthyretin, trans-oxyresveratrol | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W46
 
 | | X-ray crystal structure of V30M-TTR in complex with pterostilbene | | Descriptor: | Pterostilbene, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W43
 
 | | X-ray crystal structure of V30M-TTR in complex with piceatannol | | Descriptor: | PICEATANNOL, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W45
 
 | | X-ray crystal structure of V30M-TTR in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W47
 
 | | X-ray crystal structure of V30M-TTR in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
