8JNA
 
 | | CRAF ras-binding domain chimera, apo form | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase, CRaf | | Authors: | Kawamura, T, Kumasaka, T. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JNB
 
 | | CRAF ras-binding domain chimera, ligand complex | | Descriptor: | 2-[4-[[(2S)-1-ethanoyl-3-oxidanylidene-2H-indol-2-yl]methyl]-2-methoxy-phenoxy]ethanamide, RAF proto-oncogene serine/threonine-protein kinase, CRaf | | Authors: | Kawamura, T, Kumasaka, T. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
7QY4
 
 | | As isolated MSOX movie series dataset 5 (2 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QXK
 
 | | As isolated MSOX movie series dataset 1 (0.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QYC
 
 | | As isolated MSOX movie series dataset 20 (8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | Descriptor: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | Authors: | Fibriansah, G, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
4EFM
 
 | | Crystal structure of H-Ras G12V in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
4EFL
 
 | | Crystal structure of H-Ras WT in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
3ANX
 
 | | Crystal structure of triamine/agmatine aminopropyltransferase (SPEE) from thermus thermophilus, complexed with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, spermidine synthase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Tanaka, N, Oshima, T, Kumasaka, T. | | Deposit date: | 2010-09-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
4EFN
 
 | | Crystal structure of H-Ras Q61L in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
8IXT
 
 | |
8IXU
 
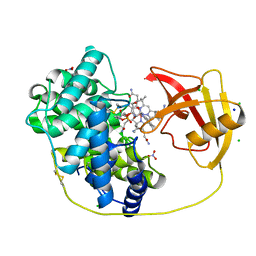 | | Rat Transcobalamin in Complex with Cobalamin | | Descriptor: | CHLORIDE ION, COBALAMIN, NITRATE ION, ... | | Authors: | Bokhove, M, Kumasaka, T. | | Deposit date: | 2023-04-03 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of the rat vitamin B 12 transporter TC and its complex with glutathionylcobalamin.
J.Biol.Chem., 300, 2024
|
|
2DCJ
 
 | | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2DCK
 
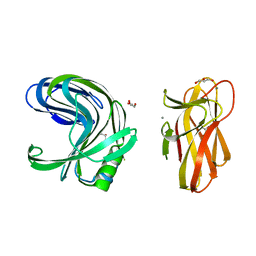 | | A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | CALCIUM ION, GLYCEROL, xylanase J | | Authors: | Fibriansah, G, Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
3ASJ
 
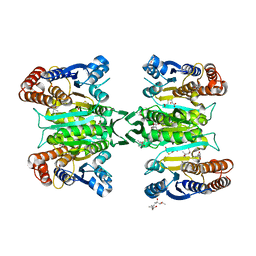 | | Crystal structure of homoisocitrate dehydrogenase in complex with a designed inhibitor | | Descriptor: | (2Z)-3-[(carboxymethyl)sulfanyl]-2-hydroxyprop-2-enoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(carboxymethyl)sulfanyl]-2-oxopropanoic acid, ... | | Authors: | Nango, E, Kumasaka, T, Eguchi, T. | | Deposit date: | 2010-12-13 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermus thermophilus homoisocitrate dehydrogenase in complex with a designed inhibitor
J.Biochem., 150, 2011
|
|
3ATN
 
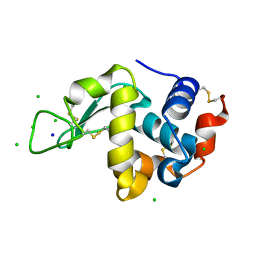 | |
3CE1
 
 | | Crystal Structure of the Cu/Zn Superoxide Dismutase from Cryptococcus liquefaciens Strain N6 | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Teh, A.H, Kanamasa, S, Kajiwara, S, Kumasaka, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Cu/Zn superoxide dismutase from the heavy-metal-tolerant yeast Cryptococcus liquefaciens strain N6.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
1ERZ
 
 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | Descriptor: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | Authors: | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | Deposit date: | 2000-04-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
3AGH
 
 | | X-ray analysis of lysozyme in the presence of 200 mM Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGI
 
 | | High resolution X-ray analysis of Arg-lysozyme complex in the presence of 500 mM Arg | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
8JOG
 
 | | solution structure of Ras Binding Domein (RBD) in C-RAF with negative allosteric modulator. | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JOF
 
 | | solution-structure of Ras Binding Domain (RBD) in C-RAF | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
1EH1
 
 | | RIBOSOME RECYCLING FACTOR FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Toyoda, T, Tin, O.F, Ito, K, Fujiwara, T, Kumasaka, T, Yamamoto, M, Garber, M.B, Nakamura, Y. | | Deposit date: | 2000-02-18 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch.
RNA, 6, 2000
|
|
5B42
 
 | | Crystal structure of the C-terminal endonuclease domain of Aquifex aeolicus MutL. | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL | | Authors: | Fukui, K, Baba, S, Kumasaka, T, Yano, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Features and Functional Dependency on beta-Clamp Define Distinct Subfamilies of Bacterial Mismatch Repair Endonuclease MutL
J.Biol.Chem., 291, 2016
|
|
6K54
 
 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
