6INX
 
 | |
4LYP
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7EEE
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with gentiobiose | | Descriptor: | GLYCEROL, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2021-03-18 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.660792 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEJ
 
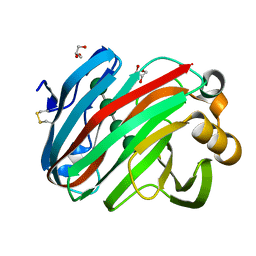 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with cellobiose | | Descriptor: | GLYCEROL, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2021-03-18 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47798049 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EE2
 
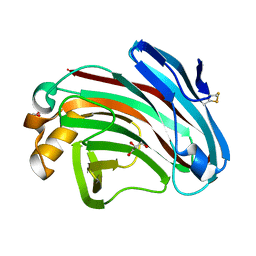 | | Structural insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.CQ31 | | Descriptor: | GLYCEROL, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | Authors: | Jiang, Z.Q, Ma, J. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37011635 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7FBT
 
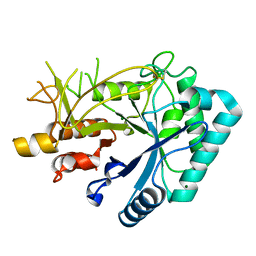 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | Descriptor: | Chitinase, MAGNESIUM ION | | Authors: | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
5YLL
 
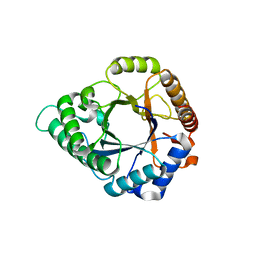 | | Structure of GH113 beta-1,4-mannanase complex with M6. | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YLI
 
 | | Complex structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YLK
 
 | | Complex structure of GH 113 family beta-1,4-mannanase with mannobiose | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase fromn Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5Z4T
 
 | | Complex structure - AxMan113A-M3 | | Descriptor: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase fromAmphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YUQ
 
 | | The high resolution structure of chitinase (RmChi1) from the thermophilic fungus Rhizomucor miehei (sp P1) | | Descriptor: | Chintase | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
6JFX
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JHF
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JEQ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with beta-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JFJ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7VGB
 
 | | Crystal structure of apo prolyl oligopeptidase from Microbulbifer arenaceous | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Huang, P, Jiang, Z.Q. | | Deposit date: | 2021-09-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | The structure and molecular dynamics of prolyl oligopeptidase from Microbulbifer arenaceous provide insights into catalytic and regulatory mechanisms.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VGC
 
 | |
