5WDR
 
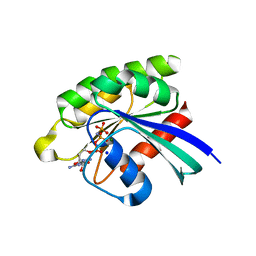 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDP
 
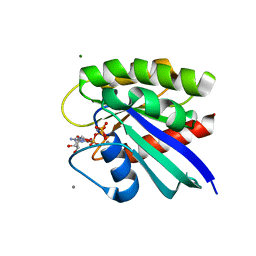 | | H-Ras mutant L120A bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
6XHZ
 
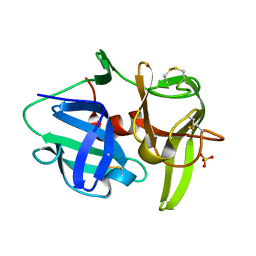 | | Alpha-lytic protease homolog N4 | | Descriptor: | N4: hypothetical protein, SULFATE ION | | Authors: | Nixon, C.F, Marqusee, S.M, Gee, C.L. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the Evolutionary History of Kinetic Stability in the alpha-Lytic Protease Family.
Biochemistry, 60, 2021
|
|
6W69
 
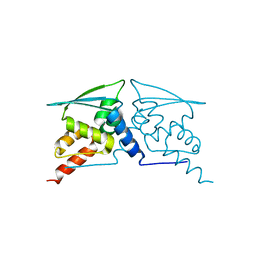 | | The structure of F64, S172A Keap1-BTB domain | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6W68
 
 | |
6W67
 
 | |
6W66
 
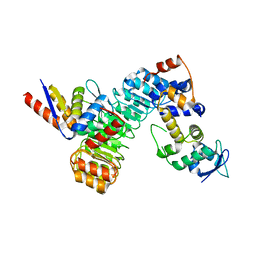 | | The structure of the F64A, S172A mutant Keap1-BTB domain in complex with SKP1-FBXL17 | | Descriptor: | F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, S-phase kinase-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6WCQ
 
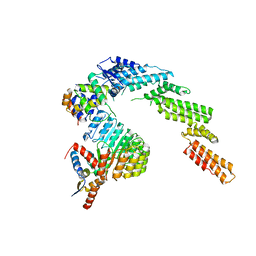 | | Structure of a substrate-bound DQC ubiquitin ligase | | Descriptor: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | Authors: | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
2OZ5
 
 | | Crystal structure of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB in complex with the specific inhibitor OMTS | | Descriptor: | Phosphotyrosine protein phosphatase ptpb, {(3-CHLOROBENZYL)[(5-{[(3,3-DIPHENYLPROPYL)AMINO]SULFONYL}-2-THIENYL)METHYL]AMINO}(OXO)ACETIC ACID | | Authors: | Grundner, C, Gee, C.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium tuberculosis Protein Tyrosine Phosphatase PtpB.
Structure, 15, 2007
|
|
5IG5
 
 | |
5IG0
 
 | | Crystal structure of S. rosetta CaMKII hub | | Descriptor: | CAMK/CAMK2 protein kinase, GLYCEROL, SULFATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG1
 
 | | Crystal structure of S. rosetta CaMKII kinase domain | | Descriptor: | CAMK/CAMK2 protein kinase, PHOSPHATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG3
 
 | | Crystal structure of the human CaMKII-alpha hub | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | McSpadden, E, Cao, Y.M, Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG4
 
 | | Crystal structure of N. vectensis CaMKII-A hub | | Descriptor: | GLYCEROL, Predicted protein | | Authors: | Bhattacharyya, M, Pappireddi, N, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
7THB
 
 | | Crystal structure of an RNA-5'/DNA-3' strand exchange junction | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*TP*AP*AP*GP*CP*AP*GP*CP*AP*TP*C)-3'), RNA (5'-R(*AP*GP*CP*UP*UP*AP*C)-3') | | Authors: | Cofsky, J.C, Knott, G.J, Gee, C.L, Doudna, J.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of an RNA/DNA strand exchange junction.
Plos One, 17, 2022
|
|
8E4T
 
 | | Crystal structure of the kinase domain of RTKC8 from the choanoflagellate Monosiga brevicollis | | Descriptor: | PHOSPHATE ION, RTKC8 Kinase domain, STAUROSPORINE | | Authors: | Bajaj, T, Gee, C.L, Kuriyan, J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the kinase domain of a receptor tyrosine kinase from a choanoflagellate, Monosiga brevicollis.
Plos One, 18, 2023
|
|
8UK9
 
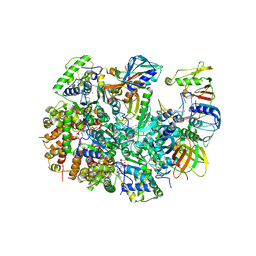 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2G8N
 
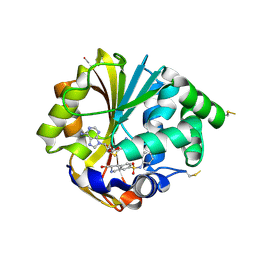 | | Structure of hPNMT with inhibitor 3-Hydroxymethyl-7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | (3R)-N-(4-CHLOROPHENYL)-3-(HYDROXYMETHYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of the Binding of 3-Fluoromethyl-7-sulfonyl-1,2,3,4-tetrahydroisoquinolines with Their Isosteric Sulfonamides to the Active Site of Phenylethanolamine N-Methyltransferase
J.Med.Chem., 49, 2006
|
|
4KH3
 
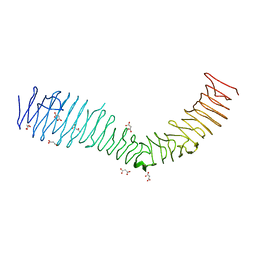 | | Structure of a bacterial self-associating protein | | Descriptor: | Antigen 43, MALONATE ION | | Authors: | Heras, B, Gee, C.L, Schembri, M.A, Totsika, M. | | Deposit date: | 2013-04-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CCD
 
 | |
3HCC
 
 | | Crystal Structure of hPNMT in Complex With anti-9-amino-5-(trifluromethyl) benzonorbornene and AdoHcy | | Descriptor: | (1S,4R,9S)-5-(trifluoromethyl)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-9-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L, Puri, M. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCB
 
 | | Crystal Structure of hPNMT in Complex With Noradrenochrome and AdoHcy | | Descriptor: | (3S)-3-hydroxy-2,3-dihydro-1H-indole-5,6-dione, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
1JGZ
 
 | | Photosynthetic Reaction Center Mutant With Tyr M 76 Replaced With Lys | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JH0
 
 | | Photosynthetic Reaction Center Mutant With Glu L 205 Replaced to Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGW
 
 | | Photosynthetic Reaction Center Mutant With Thr M 21 Replaced With Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
