4XC6
 
 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XC8
 
 | | Isobutyryl-CoA mutase fused with bound butyryl-CoA, GDP, and Mg and without cobalamin (apo-IcmF/GDP) | | Descriptor: | Butyryl Coenzyme A, GUANOSINE-5'-DIPHOSPHATE, Isobutyryl-CoA mutase fused, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XHM
 
 | |
5KDP
 
 | | E491A mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5L2R
 
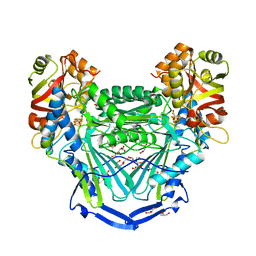 | | Crystal structure of fumarate hydratase from Leishmania major | | Descriptor: | (2S)-2-hydroxybutanedioic acid, DI(HYDROXYETHYL)ETHER, Fumarate hydratase, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8SSL
 
 | |
8USQ
 
 | |
8USP
 
 | |
6OUV
 
 | |
6OUW
 
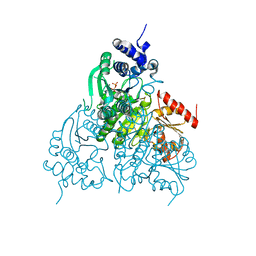 | |
3I01
 
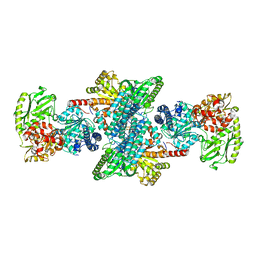 | | Native structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, water-bound C-cluster. | | Descriptor: | ACETATE ION, COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
3I04
 
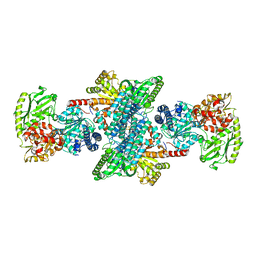 | | Cyanide-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, cyanide-bound C-cluster | | Descriptor: | ACETATE ION, COPPER (I) ION, CYANIDE ION, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
2HZV
 
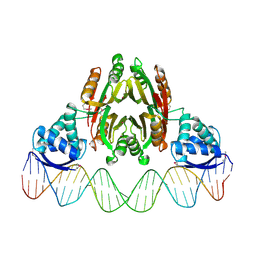 | | NikR-operator DNA complex | | Descriptor: | 5'-D(*AP*GP*TP*AP*TP*GP*AP*CP*GP*AP*AP*TP*AP*CP*TP*TP*AP*AP*AP*AP*TP*CP*GP*TP*CP*AP*TP*AP*CP*T)-3', 5'-D(*AP*GP*TP*AP*TP*GP*AP*CP*GP*AP*TP*TP*TP*TP*AP*AP*GP*TP*AP*TP*TP*CP*GP*TP*CP*AP*TP*AP*CP*T)-3', NICKEL (II) ION, ... | | Authors: | Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2006-08-09 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | NikR-operator complex structure and the mechanism of repressor activation by metal ions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HZA
 
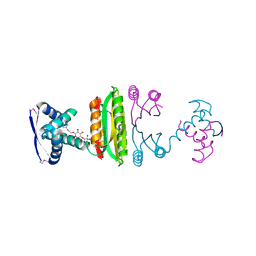 | | Nickel-bound full-length Escherichia coli NikR | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulator | | Authors: | Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NikR-operator complex structure and the mechanism of repressor activation by metal ions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4TM3
 
 | |
4TM0
 
 | |
4TLX
 
 | |
4TM1
 
 | |
4TLZ
 
 | |
4TM4
 
 | |
4TVW
 
 | | Resorufin ligase with bound resorufin-AMP analog | | Descriptor: | 5'-O-{[5-(7-hydroxy-3-oxo-3H-phenoxazin-2-yl)pentanoyl]sulfamoyl}adenosine, Lipoate-protein ligase A | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2014-06-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Computational design of a red fluorophore ligase for site-specific protein labeling in living cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TVY
 
 | | Apo resorufin ligase | | Descriptor: | 5-(3,7-dihydroxy-10H-phenoxazin-2-yl)pentanamide, Lipoate-protein ligase A | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2014-06-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Computational design of a red fluorophore ligase for site-specific protein labeling in living cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U3E
 
 | | Anaerobic ribonucleotide reductase | | Descriptor: | ACETATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-07-20 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The class III ribonucleotide reductase from Neisseria bacilliformis can utilize thioredoxin as a reductant.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3U33
 
 | |
3UBY
 
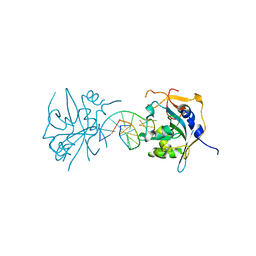 | | Crystal structure of human alklyadenine DNA glycosylase in a lower and higher-affinity complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDC)P*TP*TP*GP*CP*CP*T)-3'), DNA-3-methyladenine glycosylase | | Authors: | Setser, J.W, Lingaraju, G.M, Davis, C.A, Samson, L.D, Drennan, C.L. | | Deposit date: | 2011-10-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Searching for DNA lesions: structural evidence for lower- and higher-affinity DNA binding conformations of human alkyladenine DNA glycosylase.
Biochemistry, 51, 2012
|
|
