3V19
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
3TGL
 
 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF RHIZOMUCOR MIEHEI TRIACYLGLYCERIDE LIPASE: A CASE STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE AND MOLECULAR-MODEL REFINEMENT OF RHIZOMUCOR-MIEHEI TRIACYLGLYCERIDE LIPASE - A CASE-STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
4ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOHEXAOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOHEXAOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PMB
 
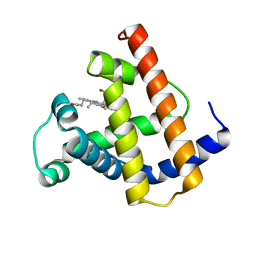 | | THE DETERMINATION OF THE CRYSTAL STRUCTURE OF RECOMBINANT PIG MYOGLOBIN BY MOLECULAR REPLACEMENT AND ITS REFINEMENT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Dodson, E.J, Dodson, G.G, Hubbard, R.E, Wilkinson, A.J. | | Deposit date: | 1989-11-27 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the crystal structure of recombinant pig myoglobin by molecular replacement and its refinement.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
3EXX
 
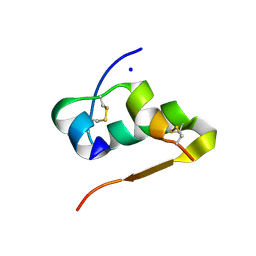 | |
1TGL
 
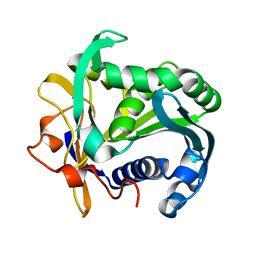 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1990-02-05 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
1BNI
 
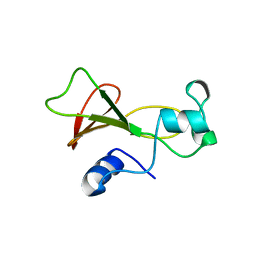 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
3W7Z
 
 | |
1EYV
 
 | | THE CRYSTAL STRUCTURE OF NUSB FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | N-UTILIZING SUBSTANCE PROTEIN B HOMOLOG, PHOSPHATE ION | | Authors: | Gopal, B, Haire, L.F, Cox, R.A, Colston, M.J, Major, S, Brannigan, J.A, Smerdon, S.J, Dodson, G.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of NusB from Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1PNL
 
 | |
1PNM
 
 | |
1PNK
 
 | |
1OS3
 
 | | Dehydrated T6 human insulin at 100 K | | Descriptor: | CHLORIDE ION, Insulin, ZINC ION | | Authors: | Smith, G.D, Blessing, R.H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lessons from an aged, dried crystal of T(6) human insulin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OS4
 
 | |
2QIU
 
 | | Structure of Human Arg-Insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Sreekanth, R, Pattabhi, V, Rajan, S.S. | | Deposit date: | 2007-07-05 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural interpretation of reduced insulin activity as seen in the crystal structure of human Arg-insulin
Biochimie, 90, 2008
|
|
3W80
 
 | |
3W7Y
 
 | |
1FN3
 
 | | CRYSTAL STRUCTURE OF NICKEL RECONSTITUTED HEMOGLOBIN-A CASE FOR PERMANENT, T-STATE HEMOGLOBIN | | Descriptor: | HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, PROTOPORPHYRIN IX CONTAINING NI(II) | | Authors: | Venkateshrao, S, Deepthi, S, Pattabhi, V, Manoharan, P.T. | | Deposit date: | 2000-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of Nickel Reconstituted Hemoglobin - A Case for Permanent, T-State Hemoglobin
CURR.SCI., 84, 2003
|
|
3INS
 
 | |
1GOY
 
 | | HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) (E.C.3.1.27.-) COMPLEXED WITH GUANOSINE-3'-PHOSPHATE (3'-GMP) | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GOV
 
 | | RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) COMPLEXED WITH SULFATE IONS | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GM9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-12 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GKF
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-13 | | Release date: | 2002-01-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM7
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
