3ZLN
 
 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 3) | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]pyridine-2-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
3ZK6
 
 | | Crystal structure of Bcl-xL in complex with inhibitor (Compound 2). | | Descriptor: | BCL-2-LIKE PROTEIN 1, N-(3-(5-(1-(2-(benzo[d]thiazol-2-yl)hydrazono)ethyl)furan-2-yl)phenylsulfonyl)-6-phenylhexanamide | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
2YQ7
 
 | | Structure of Bcl-xL bound to BimLOCK | | Descriptor: | BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
3HL5
 
 | | Crystal structure of XIAP BIR3 with CS3 | | Descriptor: | (3S)-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-3-methyl-N-(2-pyrimidin-2-ylphenyl)-L-prolinamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism of c-IAP and XIAP proteins is required for efficient induction of cell death by small-molecule IAP antagonists.
Acs Chem.Biol., 4, 2009
|
|
2YQ6
 
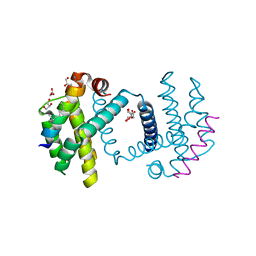 | | Structure of Bcl-xL bound to BimSAHB | | Descriptor: | BCL-2-LIKE PROTEIN 1, BIM BETA 5, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
3UW4
 
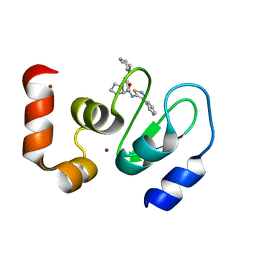 | | Crystal structure of cIAP1 BIR3 bound to GDC0152 | | Descriptor: | Baculoviral IAP repeat-containing protein 2, Baculoviral IAP repeat-containing protein 4, GDC0152, ... | | Authors: | Maurer, B, Hymowitz, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (IAP) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152).
J.Med.Chem., 55, 2012
|
|
3UW5
 
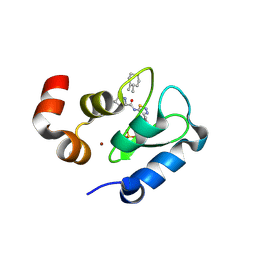 | | Crystal structure of the BIR domain of MLIAP bound to GDC0152 | | Descriptor: | Baculoviral IAP repeat-containing protein 7, Baculoviral IAP repeat-containing protein 4, GDC-0152, ... | | Authors: | Maurer, B, Hymowitz, S.G. | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (IAP) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152).
J.Med.Chem., 55, 2012
|
|
3S1K
 
 | |
1IMW
 
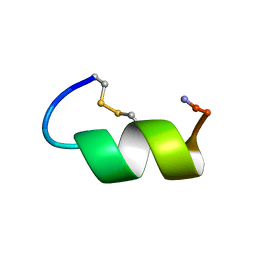 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
5DS8
 
 | |
5DTF
 
 | |
5DSC
 
 | |
5DRN
 
 | |
5DUB
 
 | |
1MFG
 
 | |
1MFL
 
 | |
