8HX6
 
 | |
8HX7
 
 | |
8HX9
 
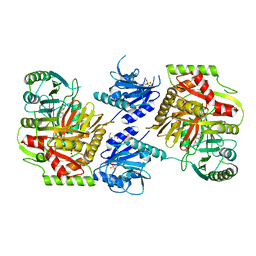 | | Crystal structure of 4-amino-4-deoxychorismate synthase from Streptomyces venezuelae with chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-amino-4-deoxychorismate synthase, FORMIC ACID, ... | | Authors: | Nakamichi, Y, Watanabe, M. | | Deposit date: | 2023-01-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the allosteric pathway of 4-amino-4-deoxychorismate synthase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8HX8
 
 | |
3VSM
 
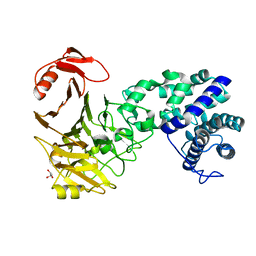 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3VSN
 
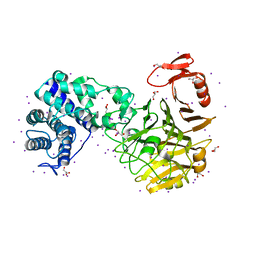 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, IODIDE ION, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
5B0U
 
 | |
8JOR
 
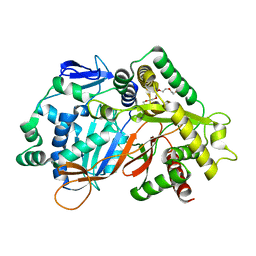 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | Descriptor: | Acyltransferase, PENTAETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
8JOS
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
1V47
 
 | | Crystal structure of ATP sulfurylase from Thermus thermophillus HB8 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase, CHLORIDE ION, ... | | Authors: | Taguchi, Y, Sugishima, M, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of a novel zinc-binding ATP sulfurylase from Thermus thermophilus HB8
Biochemistry, 43, 2004
|
|
1WUP
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81E) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
2EWI
 
 | |
2EWU
 
 | |
1WUO
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81A) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
2CDV
 
 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | Deposit date: | 1983-11-15 | | Release date: | 1984-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
3VXD
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889, SULFATE ION | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | Descriptor: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | Authors: | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7E9S
 
 | | Archaeal oligosaccharyltransferase AglB from Archaeoglobus fulgidus in complex with an inhibitory peptide and a dolichol-phosphate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, DI(HYDROXYETHYL)ETHER, Dolichyl-phosphooligosaccharide-protein glycotransferase 3, ... | | Authors: | Taguchi, Y, Hirata, K, Kohda, D. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon.
Commun Biol, 4, 2021
|
|
2YZ3
 
 | | Crystallographic Investigation of Inhibition Mode of the VIM-2 Metallo-beta-lactamase from Pseudomonas aeruginosa with Mercaptocarboxylate Inhibitor | | Descriptor: | (S)-2-(MERCAPTOMETHYL)-5-PHENYLPENTANOIC ACID, Metallo-beta-lactamase, SULFATE ION, ... | | Authors: | Yamaguchi, Y, Yamagata, Y, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2007-05-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor.
J.Med.Chem., 50, 2007
|
|
6K31
 
 | |
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
3WIW
 
 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3WVX
 
 | |
3WVY
 
 | | Structure of D48A hen egg white lysozyme in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Kawaguchi, Y, Yoneda, K, Araki, T. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The role of Asp48 in the hydrogen bonding network involving Asp52 of hen egg white lysozyme
TO BE PUBLISHED
|
|
1WIV
 
 | | solution structure of RSGI RUH-023, a UBA domain from Arabidopsis cDNA | | Descriptor: | ubiquitin-specific protease 14 | | Authors: | Higuchi, Y, Abe, T, Hirota, H, Izumi, K, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | solution structure of RSGI RUH-023, a UBA domain from Arabidopsis cDNA
To be Published
|
|
