4FIG
 
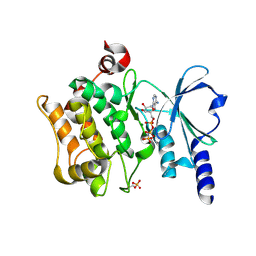 | | Catalytic domain of human PAK4 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5D68
 
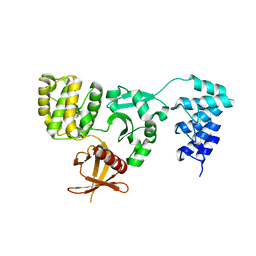 | | Crystal structure of KRIT1 ARD-FERM | | Descriptor: | Krev interaction trapped protein 1 | | Authors: | Zhang, R, Li, X, Boggon, T.J. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural analysis of the KRIT1 ankyrin repeat and FERM domains reveals a conformationally stable ARD-FERM interface.
J.Struct.Biol., 192, 2015
|
|
3L8J
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
3L8I
 
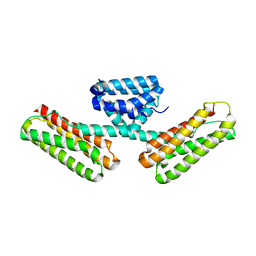 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
6PXC
 
 | |
6BHC
 
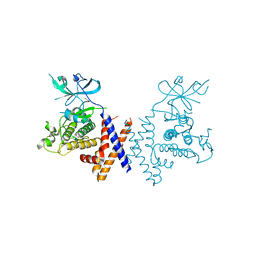 | | Crystal structure of pseduokinase PEAK1 (Sugen Kinase 269) | | Descriptor: | Pseudopodium-enriched atypical kinase 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of pseudokinase PEAK1 (Sugen kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization.
J. Biol. Chem., 293, 2018
|
|
6PXB
 
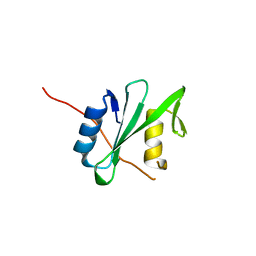 | |
6D4G
 
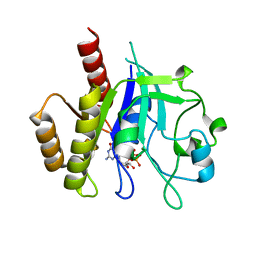 | | N-GTPase domain of p190RhoGAP-A | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 35 | | Authors: | Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2018-04-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The N-Terminal GTPase Domain of p190RhoGAP Proteins Is a PseudoGTPase.
Structure, 26, 2018
|
|
7LNY
 
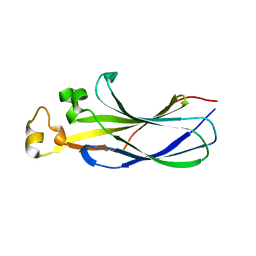 | |
7LO0
 
 | | Structure of human ASF1a in complex with a TLK2 peptide | | Descriptor: | Histone chaperone ASF1A, Serine/threonine-protein kinase tousled-like 2 | | Authors: | Simon, B, Calderwood, D, Turk, B.E, Boggon, T.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Tousled-like kinase 2 targets ASF1 histone chaperones through client mimicry.
Nat Commun, 13, 2022
|
|
5HVK
 
 | |
5HVJ
 
 | |
1OBU
 
 | | Apocrustacyanin C1 crystals grown in space and earth using vapour diffusion geometry | | Descriptor: | CRUSTACYANIN C1 SUBUNIT | | Authors: | Habash, J, Boggon, T.J, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apocrustacyanin C(1) Crystals Grown in Space and on Earth Using Vapour-Diffusion Geometry: Protein Structure Refinements and Electron-Density Map Comparisons
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OBQ
 
 | | Apocrustacyanin C1 crystals grown in space and earth using vapour diffusion geometry | | Descriptor: | CRUSTACYANIN C1 SUBUNIT | | Authors: | Habash, J, Boggon, T.J, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Apocrustacyanin C(1) Crystals Grown in Space and on Earth Using Vapour-Diffusion Geometry: Protein Structure Refinements and Electron-Density Map Comparisons
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6WAX
 
 | | C-terminal SH2 domain of p120RasGAP | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein 1, SULFATE ION | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
6WLY
 
 | |
6WLX
 
 | |
6WAY
 
 | | C-terminal SH2 domain of p120RasGAP in complex with p190RhoGAP phosphotyrosine peptide | | Descriptor: | Ras GTPase-activating protein 1, Rho GTPase-activating protein 35 | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
5U4V
 
 | |
3RQG
 
 | |
3RQF
 
 | |
5UPK
 
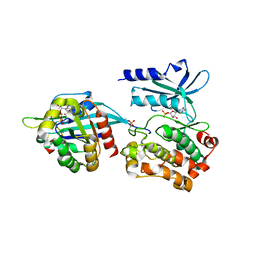 | | CDC42 binds PAK4 via an extended GTPase-effector interface - 3 peptide: PAK4cat, PAK4-N45, CDC42 | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CDC42 binds PAK4 via an extended GTPase-effector interface.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3SBE
 
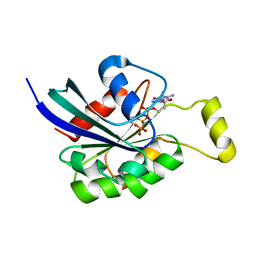 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3SBD
 
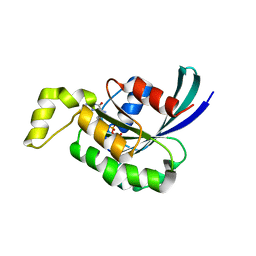 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
5VEE
 
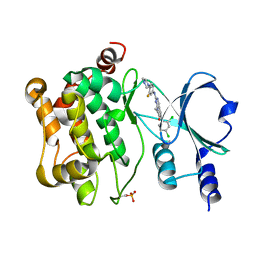 | | PAK4 kinase domain in complex with FRAX486 | | Descriptor: | 6-(2,4-dichlorophenyl)-8-ethyl-2-{[3-fluoro-4-(piperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhang, E.Y, Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PAK4 crystal structures suggest unusual kinase conformational movements.
Biochim. Biophys. Acta, 1866, 2018
|
|
