1BJT
 
 | | TOPOISOMERASE II RESIDUES 409-1201 | | Descriptor: | TOPOISOMERASE II | | Authors: | Fass, D, Bogden, C.E, Berger, J.M. | | Deposit date: | 1998-06-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands.
Nat.Struct.Biol., 6, 1999
|
|
2G2U
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
3C4O
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C4P
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
1AOL
 
 | | FRIEND MURINE LEUKEMIA VIRUS RECEPTOR-BINDING DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GP70, ZINC ION | | Authors: | Fass, D, Davey, R.A, Hamson, C.A, Kim, P.S, Cunningham, J.M, Berger, J.M. | | Deposit date: | 1997-07-08 | | Release date: | 1997-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution.
Science, 277, 1997
|
|
8B4H
 
 | | IstA transposase cleaved donor complex | | Descriptor: | DNA (55-MER) / right IS21 transposon end (insertion sequence IS5376), DNA (57-MER) / right IS21 transposon end (insertion sequence IS5376), MAGNESIUM ION, ... | | Authors: | Spinola-Amilibia, M, de la Gandara, A, Araujo-Bazan, L, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings.
Nat Commun, 14, 2023
|
|
7UGW
 
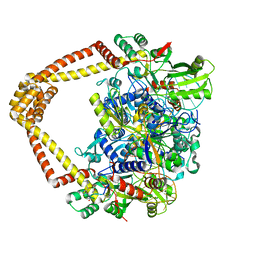 | | M. tuberculosis DNA gyrase cleavage core bound to DNA and evybactin | | Descriptor: | DNA (46-MER), DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Hauk, G, Imai, Y, Lewis, K, Berger, J.M. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis.
Nat.Chem.Biol., 18, 2022
|
|
1CKQ
 
 | |
1CL8
 
 | |
1D3Y
 
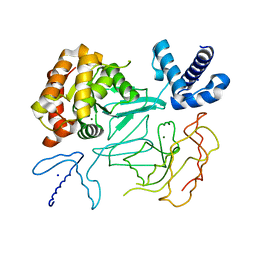 | | STRUCTURE OF THE DNA TOPOISOMERASE VI A SUBUNIT | | Descriptor: | DNA TOPOISOMERASE VI A SUBUNIT, MAGNESIUM ION, SODIUM ION | | Authors: | Nichols, M.D, DeAngelis, K.A, Keck, J.L, Berger, J.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11.
EMBO J., 18, 1999
|
|
1DDE
 
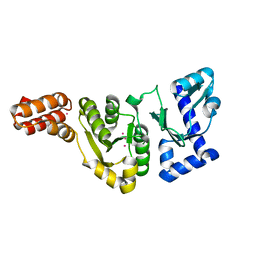 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, YTTRIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DD9
 
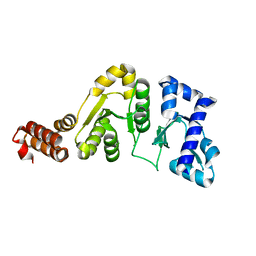 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, STRONTIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DSX
 
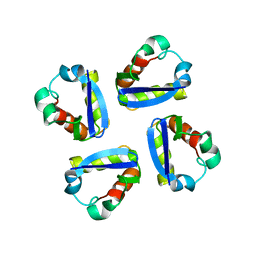 | | KV1.2 T1 DOMAIN, RESIDUES 33-119, T46V MUTANT | | Descriptor: | PROTEIN (KV1.2 VOLTAGE-GATED POTASSIUM CHANNEL) | | Authors: | Minor Jr, D.L, Lin, Y.-F, Mobley, B.C, Avelar, A, Jan, Y.N, Jan, L.Y, Berger, J.M. | | Deposit date: | 2000-01-10 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel.
Cell(Cambridge,Mass.), 102, 2000
|
|
3FK3
 
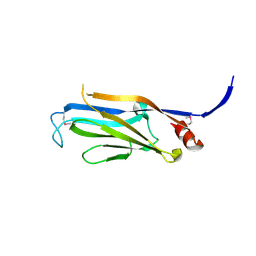 | | Structure of the Yeats Domain, Yaf9 | | Descriptor: | Protein AF-9 homolog | | Authors: | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | Deposit date: | 2008-12-15 | | Release date: | 2009-10-27 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1L8Q
 
 | |
5HE9
 
 | |
5HE8
 
 | | Bacterial initiation protein | | Descriptor: | Helicase loader, SULFATE ION | | Authors: | Hood, I.V, Berger, J.M. | | Deposit date: | 2016-01-05 | | Release date: | 2016-06-08 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bacterial initiation protein
To Be Published
|
|
1A8V
 
 | |
5BTG
 
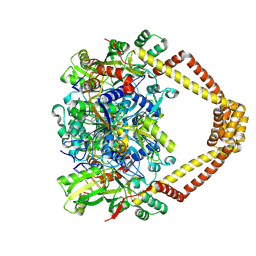 | | Crystal structure of a topoisomerase II complex | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BTL
 
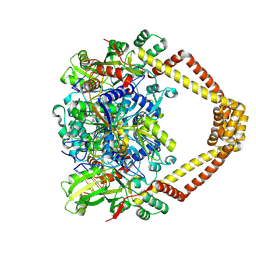 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methyl-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BTC
 
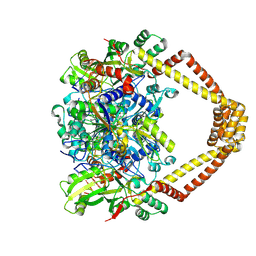 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BQ5
 
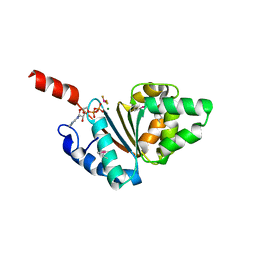 | | Crystal structure of the IstB AAA+ domain bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Insertion sequence IS5376 putative ATP-binding protein, ... | | Authors: | Arias-Palomo, E, Berger, J.M. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment.
Cell, 162, 2015
|
|
6OK8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K127L at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-04-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5CFN
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3',3' c-di-AMP, c[A(3',5')pA(3',5')p] | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
2A8V
 
 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | Descriptor: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | Authors: | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | Deposit date: | 1998-11-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
