2GIL
 
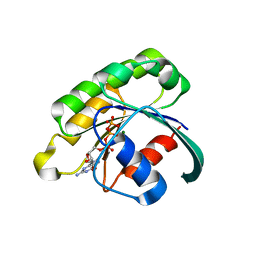 | | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Bergbrede, T, Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution
J.STRUCT.BIOL., 152, 2005
|
|
6ZQS
 
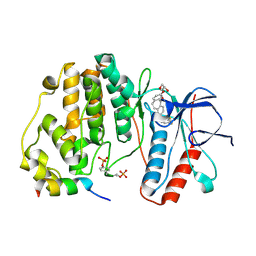 | | Crystal structure of double-phosphorylated p38alpha with ATF2(83-102) | | Descriptor: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Cyclic AMP-dependent transcription factor ATF-2, Mitogen-activated protein kinase 14 | | Authors: | Kirsch, K, Sok, P, Poti, A.L, Remenyi, A. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Nat Commun, 11, 2020
|
|
6ZR5
 
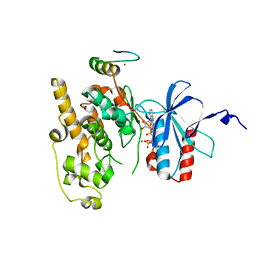 | | Crystal structure of JNK1 in complex with ATF2(19-58) | | Descriptor: | Cyclic AMP-dependent transcription factor ATF-2, MAGNESIUM ION, Mitogen-activated protein kinase 8, ... | | Authors: | Kirsch, K, Zeke, A, Remenyi, A. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Nat Commun, 11, 2020
|
|
2Y9Q
 
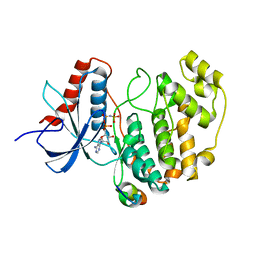 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | MAP KINASE-INTERACTING SERINE/THREONINE-PROTEIN KINASE 1, MITOGEN-ACTIVATED PROTEIN KINASE 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Barkai, T, Garai, A, Toeroe, I, Remenyi, A. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
2Y8O
 
 | | Crystal structure of human p38alpha complexed with a MAPK docking peptide | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 6, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Barkai, T, Garai, A, Toeroe, I, Remenyi, A. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
2XRW
 
 | | Linear binding motifs for JNK and for calcineurin antagonistically control the nuclear shuttling of NFAT4 | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE 8, NUCLEAR FACTOR OF ACTIVATED T-CELLS, ... | | Authors: | Barkai, T, Toeoroe, I, Garai, A, Remenyi, A. | | Deposit date: | 2010-09-23 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
2XS0
 
 | | Linear binding motifs for JNK and for calcineurin antagonistically control the nuclear shuttling of NFAT4 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 8, NUCLEAR FACTOR OF ACTIVATED T-CELLS, CYTOPLASMIC 3, ... | | Authors: | Barkai, T, Toeoroe, I, Garai, A, Remenyi, A. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
5N7F
 
 | | MAGI-1 complexed with a pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
5N7G
 
 | | MAGI-1 complexed with a synthetic pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
6TCA
 
 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
5N7D
 
 | | MAGI-1 complexed with a RSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
5CSN
 
 | | S100B-RSK1 crystal structure C | | Descriptor: | CALCIUM ION, Protein S100-B, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
5CSJ
 
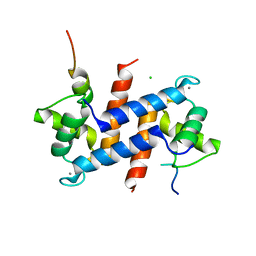 | | S100B-RSK1 crystal structure B | | Descriptor: | CALCIUM ION, CHLORIDE ION, Protein S100-B, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
5CSI
 
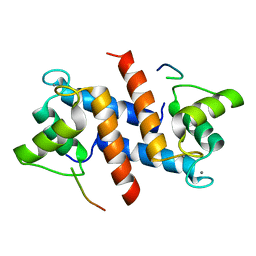 | | S100B-RSK1 crystal structure A' | | Descriptor: | CALCIUM ION, Protein S100-B, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
5CSF
 
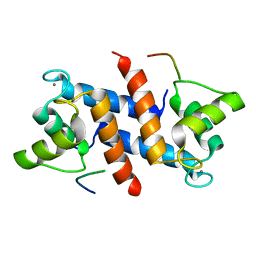 | | S100B-RSK1 crystal structure A | | Descriptor: | CALCIUM ION, Protein S100-B, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
4LH8
 
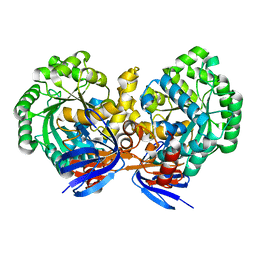 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
4FMQ
 
 | |
8OMW
 
 | | Crystal structure of the titin domain Fn3-20 | | Descriptor: | CHLORIDE ION, Titin | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OIY
 
 | |
8OSD
 
 | | Crystal structure of the titin domain Fn3-49 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OS3
 
 | | Crystal structure of the titin domain Fn3-11 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OT5
 
 | | Crystal structure of the titin domain Fn3-85 | | Descriptor: | CHLORIDE ION, SODIUM ION, Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OQ9
 
 | | Crystal structure of the titin domain Fn3-56 | | Descriptor: | CHLORIDE ION, Titin, ZINC ION | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8ORL
 
 | |
