3R1K
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis H37Rv in complex with CoA and an acetamide moiety | | Descriptor: | ACETAMIDE, COENZYME A, Enhanced intracellular survival protein | | Authors: | Biswas, T, Chen, W, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4JD6
 
 | | Crystal structure of Mycobacterium tuberculosis Eis in complex with coenzyme A and tobramycin | | Descriptor: | COENZYME A, Enhanced intracellular survival protein, TOBRAMYCIN | | Authors: | Biswas, T, Chen, W, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2013-02-23 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase eis.
Chembiochem, 14, 2013
|
|
7V0B
 
 | |
7V0D
 
 | | Crystal structure of halogenase CtcP from Kitasatospora aureofaciens | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hou, C, Tsodikov, O.V. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and complex formation of halogenase CtcP and FAD reductase CtcQ from the chlortetracycline biosynthetic pathway
To Be Published
|
|
5WMM
 
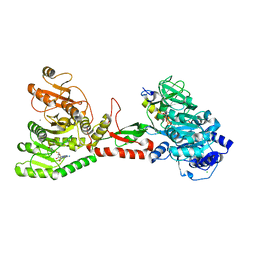 | | Crystal structure of an adenylation domain interrupted by a methylation domain (AMA4) from nonribosomal peptide synthetase TioS | | Descriptor: | (2S)-2-amino-3-methylbutanoyl (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl hydrogen (S)-phosphate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pang, A.H, Mori, S, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-07-30 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for backbone N-methylation by an interrupted adenylation domain.
Nat. Chem. Biol., 14, 2018
|
|
6B0U
 
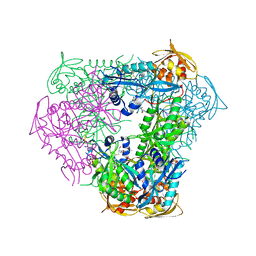 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with a Lys-containing peptide | | Descriptor: | COENZYME A, N-acetyltransferase Eis, Synthetic peptide ATKAPAKKA | | Authors: | Biswas, T, Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-09-15 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acetylation by Eis and Deacetylation by Rv1151c of Mycobacterium tuberculosis HupB: Biochemical and Structural Insight.
Biochemistry, 57, 2018
|
|
6BZQ
 
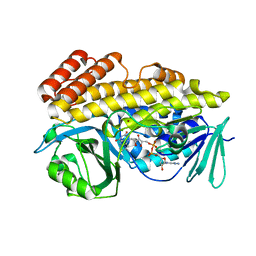 | | Crystal structure of halogenase PltM in complex with FAD | | Descriptor: | BROMIDE ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-12-25 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Unusual substrate and halide versatility of phenolic halogenase PltM.
Nat Commun, 10, 2019
|
|
6BZZ
 
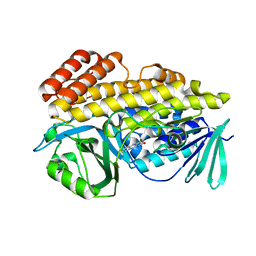 | |
6B3T
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with a 1,2,4-triazino[5,6b]indole-3-thioether inhibitor analogue 39b | | Descriptor: | 8-fluoro-5-methyl-3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-5H-[1,2,4]triazino[5,6-b]indole, COENZYME A, N-acetyltransferase Eis, ... | | Authors: | Gajadeera, C.S, Hou, C, Garneau-Tsodikova, S, Ngo, H.X, Tsodikov, O.V. | | Deposit date: | 2017-09-24 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent 1,2,4-Triazino[5,6 b]indole-3-thioether Inhibitors of the Kanamycin Resistance Enzyme Eis from Mycobacterium tuberculosis.
ACS Infect Dis, 4, 2018
|
|
6BZA
 
 | | Crystal structure of halogenase PltM in complex with phloroglucinol and FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Halogenase PltM, ... | | Authors: | Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-12-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unusual substrate and halide versatility of phenolic halogenase PltM.
Nat Commun, 10, 2019
|
|
6BZT
 
 | | Crystal structure of halogenase PltM L111Y mutant in complex with FAD | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-12-26 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unusual substrate and halide versatility of phenolic halogenase PltM.
Nat Commun, 10, 2019
|
|
7JSL
 
 | |
7JSA
 
 | |
8G7F
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and 1-hydroxypropylphosphonic acid | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
8G7G
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and (1-hydroxy-2-methylpropyl)phosphonic acid | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Metallothiol transferase FosB, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
8G7H
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and (1-hydroxypropan-2-yl)phosphonic acid | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
8G7I
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and Sulfate | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
5EBV
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor 11c and CoA | | Descriptor: | 5-(4-chlorophenyl)-~{N}-[3-(3,4-dihydro-1~{H}-isoquinolin-2-yl)propyl]-4-methyl-1,1-bis(oxidanylidene)-1,2-thiazol-3-amine, CHLORIDE ION, COENZYME A, ... | | Authors: | Gajadeera, C.S, Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2015-10-19 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent Inhibitors of Acetyltransferase Eis Overcome Kanamycin Resistance in Mycobacterium tuberculosis.
Acs Chem.Biol., 11, 2016
|
|
5EC4
 
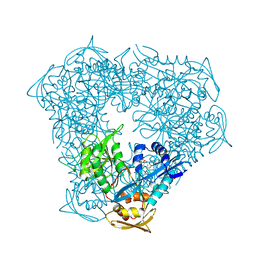 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor 13g and CoA | | Descriptor: | 5-(3-chlorophenyl)-4-methyl-~{N}-(3-morpholin-4-ylpropyl)-1,1-bis(oxidanylidene)-1,2-thiazol-3-amine, COENZYME A, Enhanced intracellular survival protein | | Authors: | Gajadeera, C.S, Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2015-10-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent Inhibitors of Acetyltransferase Eis Overcome Kanamycin Resistance in Mycobacterium tuberculosis.
Acs Chem.Biol., 11, 2016
|
|
3LT3
 
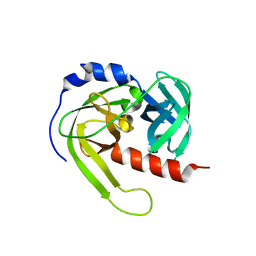 | | Crystal structure of Rv3671c from M. tuberculosis H37Rv, Ser343Ala mutant, inactive form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2010-02-14 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
3K6Y
 
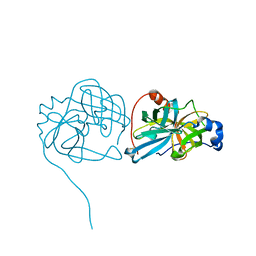 | | Crystal structure of Rv3671c protease from M. tuberculosis, active form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2009-10-10 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
6X10
 
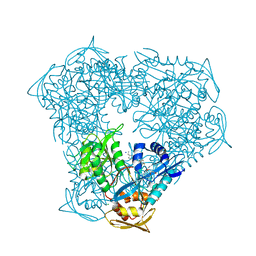 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with haloperidol | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-17 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
6X6G
 
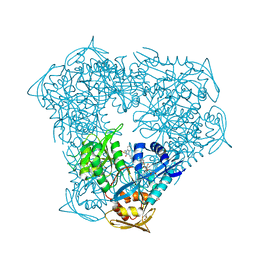 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with droperidol | | Descriptor: | 3-[1-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-2,3,4,5-tetrahydropyridin-4-yl]-1~{H}-benzimidazol-2-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
6X6I
 
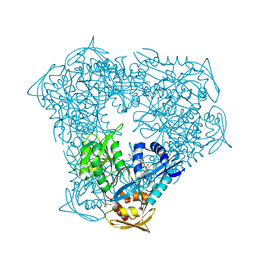 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT543 | | Descriptor: | 4-(4-benzyl-4-hydroxypiperidin-1-yl)-1-(4-fluorophenyl)butan-1-one, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
6X6Y
 
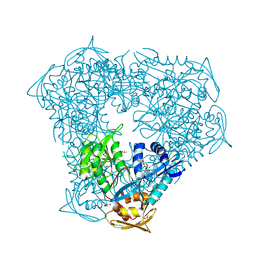 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1264 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-05-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design of haloperidol analogues as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis to overcome kanamycin resistance
Rsc Med Chem, 12, 2021
|
|
