5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
7FAT
 
 | | Structure Determination of the RBD-NB1A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | Authors: | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
4MTI
 
 | | Crystal structure of cIAP1 BIR3 bound to T3258042 | | Descriptor: | (3S,8aR)-2-[(2S)-2-cyclohexyl-2-{[(2S)-2-(methylamino)butanoyl]amino}acetyl]-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Snell, G.P, Dougan, D.R. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, synthesis, and biological activities of novel hexahydropyrazino[1,2-a]indole derivatives as potent inhibitors of apoptosis (IAP) proteins antagonists with improved membrane permeability across MDR1 expressing cells.
Bioorg.Med.Chem., 21, 2013
|
|
4MU7
 
 | | Crystal structure of cIAP1 BIR3 bound to T3450325 | | Descriptor: | (3S,10aS)-2-[(2S)-2-cyclohexyl-2-{[(2S)-2-(methylamino)butanoyl]amino}acetyl]-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-1,2,3,4,10,10a-hexahydropyrazino[1,2-a]indole-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Snell, G.P, Dougan, D.R. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design, synthesis, and biological activities of novel hexahydropyrazino[1,2-a]indole derivatives as potent inhibitors of apoptosis (IAP) proteins antagonists with improved membrane permeability across MDR1 expressing cells.
Bioorg.Med.Chem., 21, 2013
|
|
3CM5
 
 | |
4FI9
 
 | | Structure of human SUN-KASH complex | | Descriptor: | Nesprin-2, SUN domain-containing protein 2 | | Authors: | Wang, W.J, Shi, Z.B. | | Deposit date: | 2012-06-08 | | Release date: | 2012-07-18 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
8SE6
 
 | | NKG2D complexed with inhibitor 36 | | Descriptor: | N-[(1S)-2-(dimethylamino)-2-oxo-1-{3-[3-(2,2,2-trifluoroethyl)azetidin-1-yl]phenyl}ethyl]-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein, TETRAETHYLENE GLYCOL, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Development of small molecule inhibitors of natural killer group 2D receptor (NKG2D).
Bioorg.Med.Chem.Lett., 96, 2023
|
|
8SE5
 
 | | NKG2D complexed with inhibitor 14 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-[(1R,5S,6S)-6-(hydroxymethyl)-3-azabicyclo[3.1.0]hexan-3-yl]-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Development of small molecule inhibitors of natural killer group 2D receptor (NKG2D).
Bioorg.Med.Chem.Lett., 96, 2023
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
8Y3W
 
 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (same side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y13
 
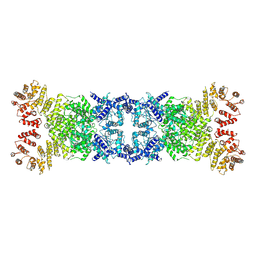 | | Cryo-EM structure of anti-phage defense associated DSR2 tetramer (H171A) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Li, F.X, Shi, Z.B, Wang, R.W, Xu, Q, Yang, R, Wu, Z.X. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y34
 
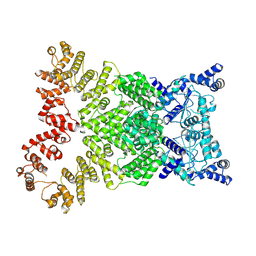 | | Cryo-EM structure of anti-phage defense associated DSR2 (H171A) (map2) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-28 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3M
 
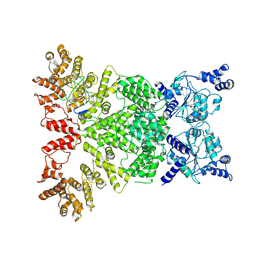 | | Cryo-EM structure of DSR2-DSAD1 complex (cross-linked) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3Y
 
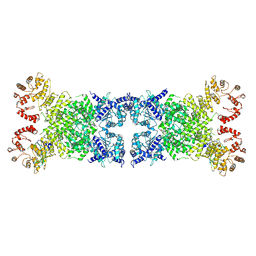 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (opposite side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8EA8
 
 | | NKG2D complexed with inhibitor 4a | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA7
 
 | | NKG2D complexed with inhibitor 3g | | Descriptor: | (4M)-N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-(1-methyl-1H-pyrazol-5-yl)-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, DI(HYDROXYETHYL)ETHER, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA5
 
 | | NKG2D complexed with inhibitor 1a | | Descriptor: | (3S,5aS,8aR)-3-benzyl-6-[(3,5-dichlorophenyl)methyl]-1,4-dimethyloctahydropyrrolo[3,2-e][1,4]diazepine-2,5-dione, NKG2-D type II integral membrane protein, TRIETHYLENE GLYCOL | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA9
 
 | | NKG2D complexed with inhibitor 4d | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-1-[3,5-bis(trifluoromethyl)phenyl]-2-(dimethylamino)-2-oxoethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAB
 
 | | NKG2D complexed with inhibitor 4f | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-2-oxo-1-[3-(trifluoromethyl)phenyl]-2-({4-[4-(trifluoromethyl)phenyl]pyridin-3-yl}amino)ethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAA
 
 | | NKG2D complexed with inhibitor 4e | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{(1S)-2-(dimethylamino)-1-[3-methyl-5-(trifluoromethyl)phenyl]-2-oxoethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA6
 
 | | NKG2D complexed with inhibitor 3e | | Descriptor: | N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7VFB
 
 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFA
 
 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4GEH
 
 | |
