2RLZ
 
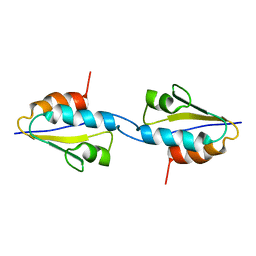 | | Solid-State MAS NMR structure of the dimer Crh | | Descriptor: | HPr-like protein crh | | Authors: | Loquet, A, Bardiaux, B, Gardiennet, C, Blanchet, C, Baldus, M, Nilges, M, Malliavin, T, Bockmann, A. | | Deposit date: | 2007-09-04 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | 3D structure determination of the Crh protein from highly ambiguous solid-state NMR restraints.
J.Am.Chem.Soc., 130, 2008
|
|
2BJX
 
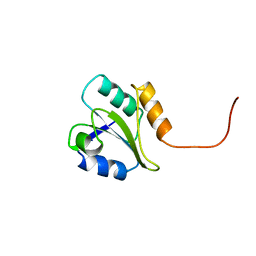 | | PROTEIN DISULFIDE ISOMERASE | | Descriptor: | PROTEIN (PROTEIN DISULFIDE ISOMERASE) | | Authors: | Kemmink, J, Dijkstra, K, Mariani, M, Scheek, R.M, Penka, E, Nilges, M, Darby, N.J. | | Deposit date: | 1999-01-30 | | Release date: | 1999-02-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure in solution of the b domain of protein disulfide isomerase.
J.Biomol.NMR, 13, 1999
|
|
1KKS
 
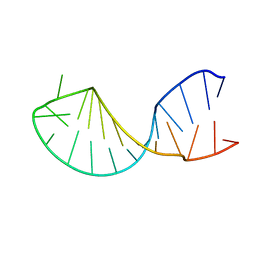 | | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression | | Descriptor: | 5'-R(*GP*GP*AP*AP*GP*GP*CP*CP*CP*UP*UP*UP*UP*CP*AP*GP*GP*GP*CP*CP*AP*CP*CP*C)-3' | | Authors: | Zanier, K, Luyten, I, Crombie, C, Muller, B, Schuemperli, D, Linge, J.P, Nilges, M, Sattler, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression.
RNA, 8, 2002
|
|
6GV9
 
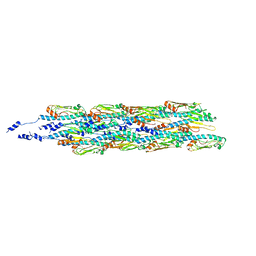 | | Structure of the type IV pilus from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Bardiaux, B, Amorim, G.C, Luna-Rico, A, Zheng, W, Guilvout, I, Jollivet, C, Nilges, M, Egelman, E, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
1M7L
 
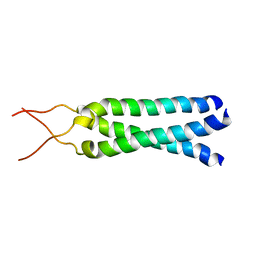 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
1BPV
 
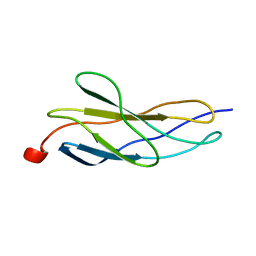 | |
1JUN
 
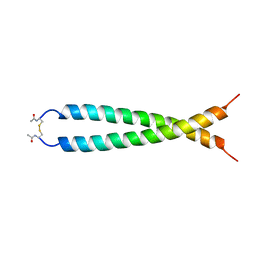 | |
1QXN
 
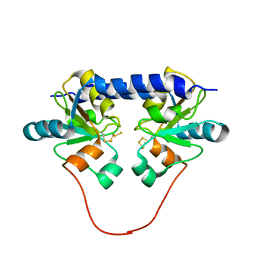 | | Solution Structure of the 30 kDa Polysulfide-sulfur Transferase Homodimer from Wolinella Succinogenes | | Descriptor: | PENTASULFIDE-SULFUR, sulfide dehydrogenase | | Authors: | Lin, Y.J, Dancea, F, Loehr, F, Klimmek, O, Pfeiffer-Marek, S, Nilges, M, Wienk, H, Kroeger, A, Rueterjans, H. | | Deposit date: | 2003-09-08 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 30 kDa Polysulfide-Sulfur Transferase Homodimer from Wolinella succinogenes
Biochemistry, 43, 2004
|
|
2AIY
 
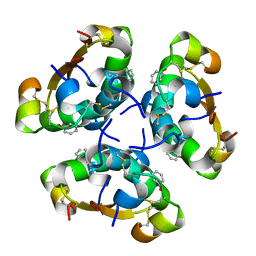 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 20 STRUCTURES | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-28 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
1PFS
 
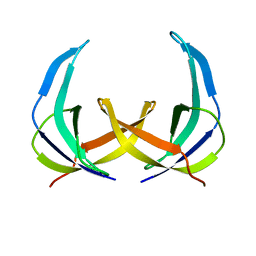 | | SOLUTION NMR STRUCTURE OF THE SINGLE-STRANDED DNA BINDING PROTEIN OF THE FILAMENTOUS PSEUDOMONAS PHAGE PF3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PF3 SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Folmer, R.H.A, Nilges, M, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1996-08-03 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the single-stranded DNA binding protein of the filamentous Pseudomonas phage Pf3: similarity to other proteins binding to single-stranded nucleic acids.
EMBO J., 14, 1995
|
|
1VIG
 
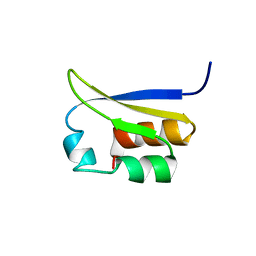 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VIH
 
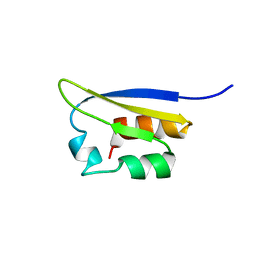 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
2KUB
 
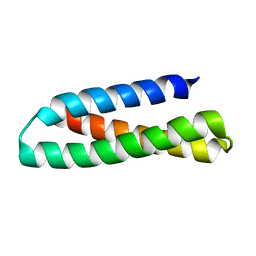 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
2J0E
 
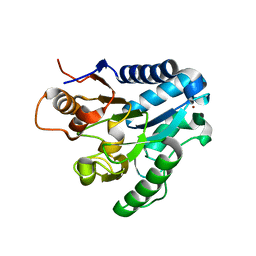 | | Three dimensional structure and catalytic mechanism of 6- phosphogluconolactonase from Trypanosoma brucei | | Descriptor: | 6-PHOSPHOGLUCONOLACTONASE, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Delarue, M, Duclert-Savatier, N, Miclet, E, Haouz, A, Giganti, D, Ouazzani, J, Lopez, P, Nilges, M, Stoven, V. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Dimensional Structure and Implications for the Catalytic Mechanism of 6-Phosphogluconolactonase from Trypanosoma Brucei.
J.Mol.Biol., 366, 2007
|
|
2GVB
 
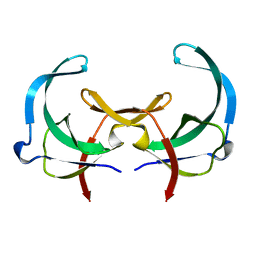 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
2GVA
 
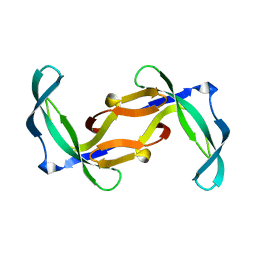 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
7PKI
 
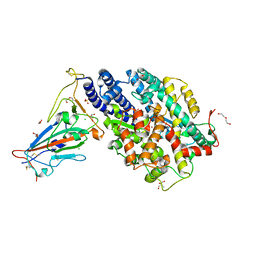 | |
1F5J
 
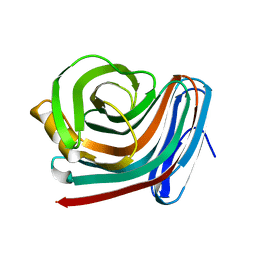 | | CRYSTAL STRUCTURE OF XYNB, A HIGHLY THERMOSTABLE BETA-1,4-XYLANASE FROM DICTYOGLOMUS THERMOPHILUM RT46B.1, AT 1.8 A RESOLUTION | | Descriptor: | BETA-1,4-XYLANASE, SULFATE ION | | Authors: | McCarthy, A.A, Baker, E.N. | | Deposit date: | 2000-07-26 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of XynB, a highly thermostable beta-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1, at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FL1
 
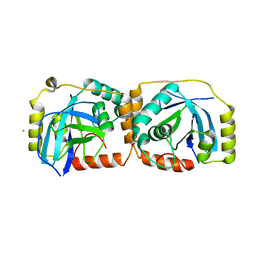 | | KSHV PROTEASE | | Descriptor: | POTASSIUM ION, PROTEASE | | Authors: | Reiling, K.K, Pray, T.R, Craik, C.S, Stroud, R.M. | | Deposit date: | 2000-08-11 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements.
Biochemistry, 39, 2000
|
|
3E7F
 
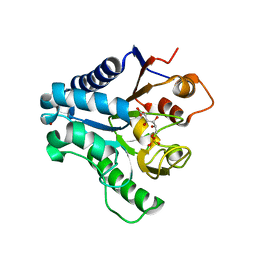 | | Crystal structure of 6-phosphogluconolactonase from Trypanosoma brucei complexed with 6-phosphogluconic acid | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconolactonase, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-18 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
3EB9
 
 | | Crystal structure of 6-phosphogluconolactonase from trypanosoma brucei complexed with citrate | | Descriptor: | 6-phosphogluconolactonase, CITRATE ANION, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-27 | | Release date: | 2009-05-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
8C67
 
 | | Crystal structure of Ab25 Fab | | Descriptor: | antibody 25 heavy chain, antibody 25 light chain | | Authors: | Nyblom, M, Izadi, A, Tang, D, Bahnan, W, Happonen, L, Malmstroem, J, Shannon, O, Malmstroem, L, Nordenfelt, P. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering of IgG1 hinge to the flexible IgG3 hinge enhances immune defense against streptococci
To Be Published
|
|
1WCO
 
 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BTN
 
 | | STRUCTURE OF THE BINDING SITE FOR INOSITOL PHOSPHATES IN A PH DOMAIN | | Descriptor: | BETA-SPECTRIN, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE | | Authors: | Wilmanns, M, Hyvoenen, M, Saraste, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the binding site for inositol phosphates in a PH domain.
EMBO J., 14, 1995
|
|
