6AA3
 
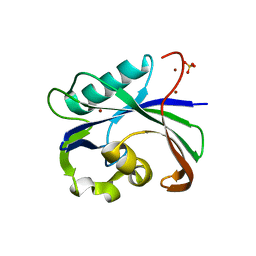 | | Crystal structure of MTH1 in apo form (cocktail No. 1) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, ZINC ION | | Authors: | Yokoyama, T, Kitakami, R, Mizuguchi, M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a new class of MTH1 inhibitor by X-ray crystallographic screening.
Eur J Med Chem, 167, 2019
|
|
2JX8
 
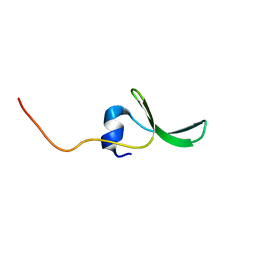 | | Solution structure of hPCIF1 WW domain | | Descriptor: | Phosphorylated CTD-interacting factor 1 | | Authors: | Kouno, T, Iwamoto, Y, Hirose, Y, Aizawa, T, Demura, M, Kawano, K, Ohkuma, Y, Mizuguchi, M. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, and 15N resonance assignments of hPCIF1 WW domain
To be Published
|
|
6AA5
 
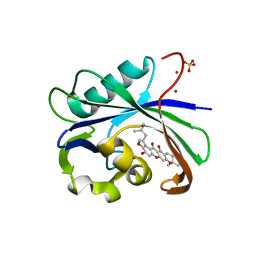 | | Crystal structure of MTH1 in complex with 3-isomangostin | | Descriptor: | 5,9-dihydroxy-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H,6H-pyrano[3,2-b]xanthen-6-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, ... | | Authors: | Yokoyama, T, Kitakami, R, Mizuguchi, M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Discovery of a new class of MTH1 inhibitor by X-ray crystallographic screening.
Eur J Med Chem, 167, 2019
|
|
6AAR
 
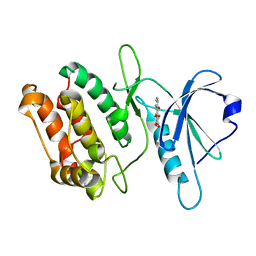 | | Crystal structure of DAPK1 in complex with purpurin | | Descriptor: | Death-associated protein kinase 1, Purpurin | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic analyses of interactions between death-associated protein kinase 1 and anthraquinones.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6AJV
 
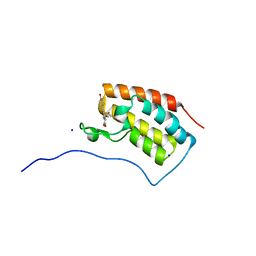 | | Crystal structure of BRD4 in complex with isoliquiritigenin and DMSO (Cocktail No. 3) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
5CG5
 
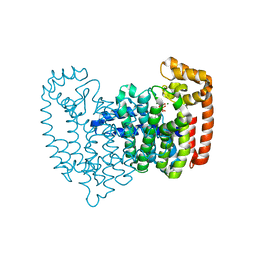 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.402 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
5CG6
 
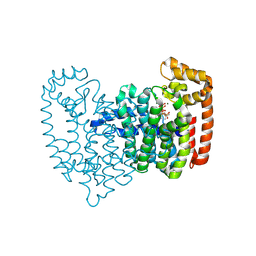 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate and isopentenyl pyrophosphate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
6AJX
 
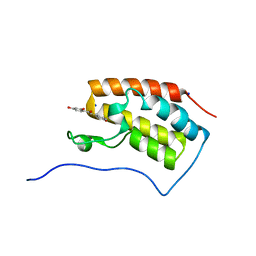 | | Crystal structure of BRD4 in complex with isoliquiritigenin in the absence of DMSO | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJY
 
 | | Crystal structure of BRD4 in complex with 2',4'-dihydroxy-2-methoxychalcone | | Descriptor: | 2',4'-dihydroxy-2-methoxychalcone, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJZ
 
 | | Joint nentron and X-ray structure of BRD4 in complex with colchicin | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, SODIUM ION | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (1.301 Å), X-RAY DIFFRACTION | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJW
 
 | | Crystal structure of BRD4 in complex with DMSO (Cocktail No. 4) | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
1IRR
 
 | | Solution structure of paralytic peptide of the silkworm, Bombyx mori | | Descriptor: | paralytic peptide | | Authors: | Miura, K, Kamimura, M, Aizawa, T, Kiuchi, M, Hayakawa, Y, Mizuguchi, M, Kawano, K. | | Deposit date: | 2001-10-23 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of paralytic peptide of silkworm, Bombyx mori
peptides, 23, 2002
|
|
3U2J
 
 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3U2I
 
 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
2KOZ
 
 | |
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
2KP0
 
 | |
1V50
 
 | | Solution structure of phosphorylated N-terminal fragment of S100C/A11 protein | | Descriptor: | Calgizzarin | | Authors: | Kouno, T, Mizuguchi, M, Sakaguchi, M, Makino, E, Huh, N, Kawano, K. | | Deposit date: | 2003-11-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Study on structure-activity relationship between the N-terminal region of S100C protein and its function
Peptide Science, 40, 2003
|
|
1V4Z
 
 | | Solution structure of the N-terminal fragment of S100C/A11 protein | | Descriptor: | Calgizzarin | | Authors: | Kouno, T, Mizuguchi, M, Sakaguchi, M, Makino, E, Huh, N, Kawano, K. | | Deposit date: | 2003-11-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Study on structure-activity relationship between the N-terminal region of S100C protein and its function
Peptide Science, 40, 2003
|
|
1V49
 
 | | Solution structure of microtubule-associated protein light chain-3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kouno, T, Mizuguchi, M, Tanida, I, Ueno, T, Kominami, E, Kawano, K. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains.
J.Biol.Chem., 280, 2005
|
|
5FVK
 
 | | Crystal structure of Vps4-Vfa1 complex from S.cerevisiae at 1.66 A resolution. | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4, VPS4-ASSOCIATED PROTEIN 1 | | Authors: | Kojima, R, Obita, T, Onoue, K, Mizuguchi, M. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Structural Fine-Tuning of Mit Interacting Motif 2 (Mim2) and Allosteric Regulation of Escrt-III by Vps4 in Yeast.
J.Mol.Biol., 428, 2016
|
|
5FVL
 
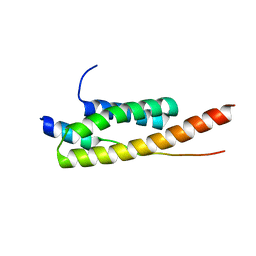 | | Crystal structure of Vps4-Vps20 complex from S.cerevisiae | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 20, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4 | | Authors: | Kojima, R, Obita, T, Onoue, K, Mizuguchi, M. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Structural Fine-Tuning of Mit Interacting Motif 2 (Mim2) and Allosteric Regulation of Escrt-III by Vps4 in Yeast.
J.Mol.Biol., 428, 2016
|
|
5H0Z
 
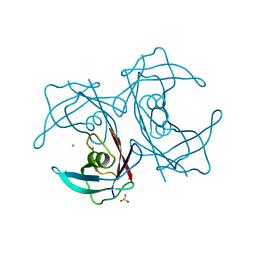 | | Crystal structure of P113A mutated human transthyretin | | Descriptor: | CHLORIDE ION, SULFATE ION, Transthyretin | | Authors: | Yokoyama, T, Hanawa, Y, Obita, T, Mizuguchi, M. | | Deposit date: | 2016-10-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | Stability and crystal structures of His88 mutant human transthyretins
FEBS Lett., 591, 2017
|
|
5H0V
 
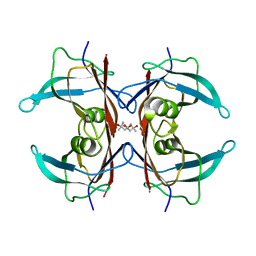 | | Crystal structure of H88A mutated human transthyretin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Transthyretin | | Authors: | Yokoyama, T, Hanawa, Y, Obita, T, Mizuguchi, M. | | Deposit date: | 2016-10-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Stability and crystal structures of His88 mutant human transthyretins
FEBS Lett., 591, 2017
|
|
5H0X
 
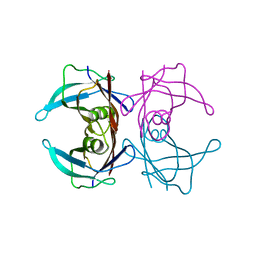 | |
