6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYJ
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6UIB
 
 | | Crystal structure of IL23 bound to peptide 23-652 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Durbin, J.D, Wang, J, Afshar, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Integration of phage and yeast display platforms: A reliable and cost effective approach for binning of peptides as displayed on-phage.
Plos One, 15, 2020
|
|
4MFI
 
 | | Crystal structure of Mycobacterium tuberculosis UgpB | | Descriptor: | Sn-glycerol-3-phosphate ABC transporter substrate-binding protein UspB | | Authors: | Jiang, D, Bartlam, M, Rao, Z. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine
Febs J., 281, 2014
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | Descriptor: | GLYCEROL, L-threonine aldolase | | Authors: | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5ZII
 
 | |
5ZKZ
 
 | |
5ZIW
 
 | |
3HGG
 
 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
3IBD
 
 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
3I7G
 
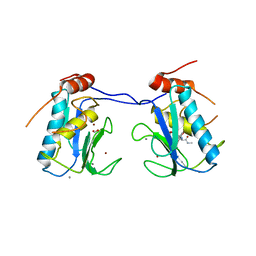 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | 5-(4-chlorophenyl)-N-[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]furan-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3I7I
 
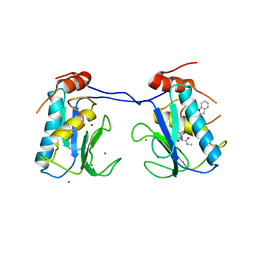 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N-[4-(5-{[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]carbamoyl}furan-2-yl)phenyl]-1-benzofuran-2-carboxamide, ... | | Authors: | Farrow, N.A, Margarit, S.M. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3UAS
 
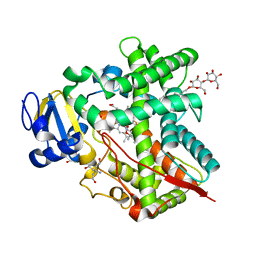 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
3HGY
 
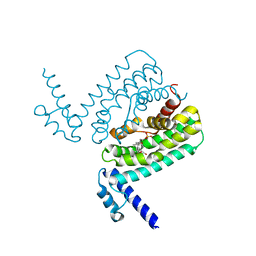 | | Crystal Structure of CmeR Bound to Taurocholic Acid | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: crystal structures of CmeR-bile acid complexes
To be Published
|
|
5T0O
 
 | | Crystal Structure of a membrane protein | | Descriptor: | CmeB | | Authors: | Su, C.-C, Yu, E.W. | | Deposit date: | 2016-08-16 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump.
Nat Commun, 8, 2017
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
3JBZ
 
 | |
5DGB
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA EXTENDING AN 1,N6-ETHENODEOXYADENOSINE : dA PAIR BY INSERTING dTMPNPP OPPOSITE TEMPLATE dA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), DNA (5'-D(*CP*AP*TP*AP*(EDA)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5E58
 
 | | Crystal Structure Of Cytochrome P450 2B35 from Desert Woodrat Neotoma Lepida in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 family 2 subfamily B, ... | | Authors: | Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Function Analysis of Mammalian CYP2B Enzymes Using 7-Substituted Coumarin Derivatives as Probes: Utility of Crystal Structures and Molecular Modeling in Understanding Xenobiotic Metabolism.
Mol.Pharmacol., 89, 2016
|
|
5DGA
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA EXTENDING AN 1,N6-ETHENODEOXYADENOSINE : dT PAIR BY INSERTING dTMPNPP OPPOSITE TEMPLATE dA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*AP*(EDA)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5DG8
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA INSERTING dAMPNPP ACROSS A DNA TEMPLATE CONTAINING 1,N6-ETHENODEOXYADENOSINE LESION | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(EDA)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5DG7
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA INSERTING dTTP ACROSS A DNA TEMPLATE CONTAINING 1,N6-ETHENODEOXYADENOSINE LESION | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(EDA)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
8TTZ
 
 | | Crystal structure of monkey TLR7 ectodomain with compound 20 | | Descriptor: | (3S)-3-({5-amino-1-[(2-methoxy-4-{[(oxan-4-yl)amino]methyl}phenyl)methyl]-1H-pyrazolo[4,3-d]pyrimidin-7-yl}amino)hexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Critton, D.A. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of Novel TLR7 Agonists as Systemic Agent for Combination With aPD1 for Use in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
