8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
4KP3
 
 | | Crystal Structure of MyoVa-GTD in Complex with Two Cargos | | Descriptor: | Melanophilin, RILP-like protein 2, Unconventional myosin-Va | | Authors: | Wei, Z, Liu, X, Yu, C, Zhang, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K7E
 
 | | Crystal structure of Junin virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Zhang, Y.J, Li, L, Liu, X, Dong, S.S, Wang, W.M, Huo, T, Rao, Z.H, Yang, C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Junin virus nucleoprotein
J.Gen.Virol., 94, 2013
|
|
3URG
 
 | | The crystal structure of Anabaena CcbP | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Alr1010 protein, SODIUM ION | | Authors: | Fan, X.X, Liu, X, Su, X.D. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Ellman's reagent in promoting crystallization and structure determination of Anabaena CcbP.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2L8S
 
 | |
3WB8
 
 | | Crystal Structure of MyoVa-GTD | | Descriptor: | 1,2-ETHANEDIOL, Unconventional myosin-Va | | Authors: | Wei, Z, Liu, X, Yu, C, Zhang, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GO1
 
 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | Descriptor: | GLYCEROL, Transcriptional regulator LsrR | | Authors: | Wu, M, Tao, Y, Liu, X, Zang, J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
5ZHP
 
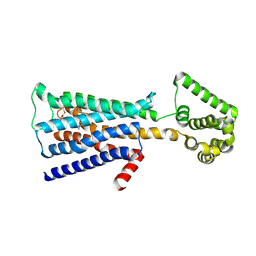 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A6O
 
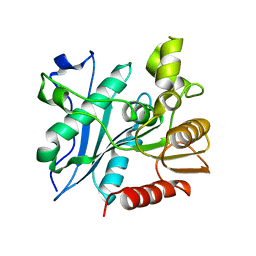 | | Crystal structure of acetyl ester-xyloside bifunctional hydrolase from Caldicellulosiruptor lactoaceticus | | Descriptor: | Esterase/lipase-like protein | | Authors: | Cao, H, Huang, Y, Sun, L.C, Liu, X, Liu, T.F, Wang, F.Z, Xin, F.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Dual-Substrate Recognition and Catalytic Mechanisms of a Bifunctional Acetyl Ester-Xyloside Hydrolase from Caldicellulosiruptor lactoaceticus.
Acs Catalysis, 9, 2019
|
|
6IY3
 
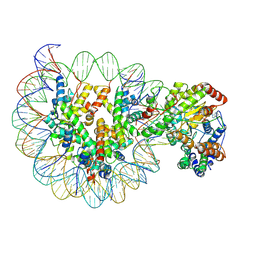 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6IY2
 
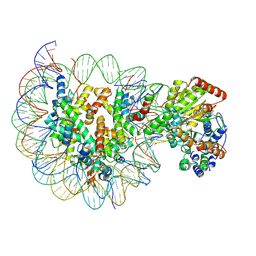 | | Structure of Snf2-MMTV-A nucleosome complex at shl2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3V
 
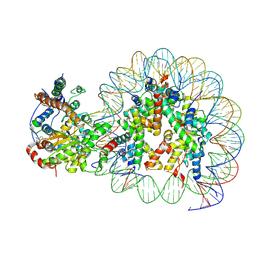 | | Structure of Snf2-nucleosome complex at shl-2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3U
 
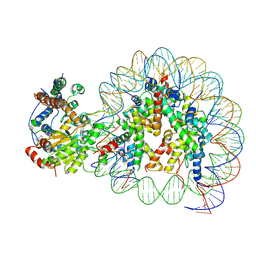 | | Structure of Snf2-nucleosome complex at shl2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3O
 
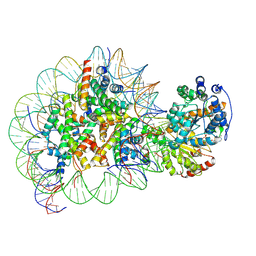 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
5Z3L
 
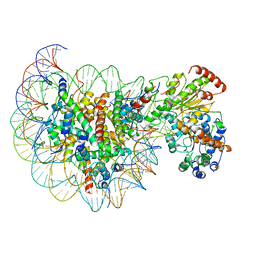 | | Structure of Snf2-nucleosome complex in apo state | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5YK9
 
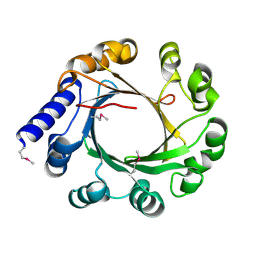 | |
5ZCJ
 
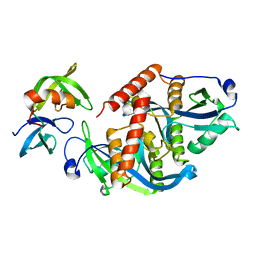 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
5ZMY
 
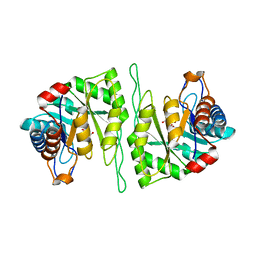 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, D(-)-TARTARIC ACID, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
5ZMU
 
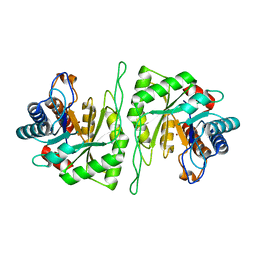 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
6JBZ
 
 | |
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
