1MRC
 
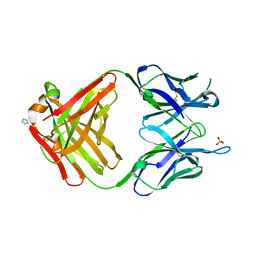 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
4EO8
 
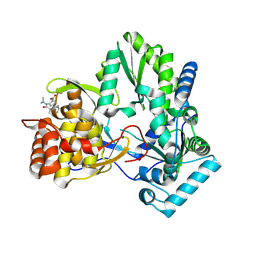 | | HCV NS5B polymerase inhibitors: Tri-substituted acylhydrazines as tertiary amide bioisosteres | | Descriptor: | 5-(3,3-dimethylbut-1-yn-1-yl)-3-{2,2-dimethyl-1-[(trans-4-methylcyclohexyl)carbonyl]hydrazinyl}thiophene-2-carboxylic acid, RNA-directed RNA polymerase | | Authors: | Appleby, T.C, Canales, E, Watkins, W.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2KCC
 
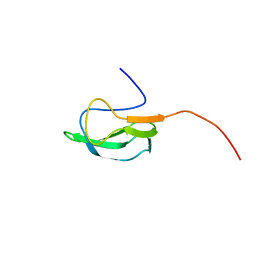 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
1OB0
 
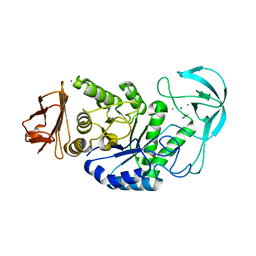 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
6BVG
 
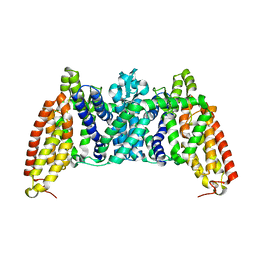 | | Crystal structure of bcMalT T280C-E54C crosslinked by divalent mercury | | Descriptor: | MERCURY (II) ION, Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ren, Z, Zhou, M. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an EIIC sugar transporter trapped in an inward-facing conformation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BL5
 
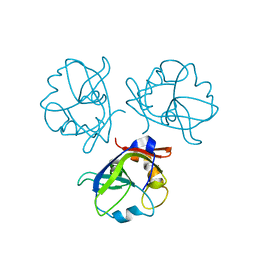 | | Head decoration protein from the hyperthermophilic phage P74-26 | | Descriptor: | Head decoration protein | | Authors: | Stone, N.P, Hilbert, B.J, Hidalgo, D, Halloran, K.T, Kelch, B.A. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Hyperthermophilic Phage Decoration Protein Suggests Common Evolutionary Origin with Herpesvirus Triplex Proteins and an Anti-CRISPR Protein.
Structure, 26, 2018
|
|
5FIG
 
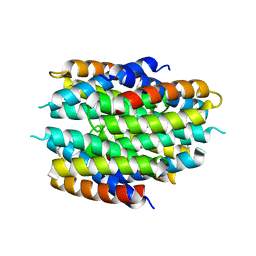 | | APO-CSP3 (COPPER STORAGE PROTEIN 3) FROM BACILLUS SUBTILIS | | Descriptor: | CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity.
Sci Rep, 6, 2016
|
|
6ISG
 
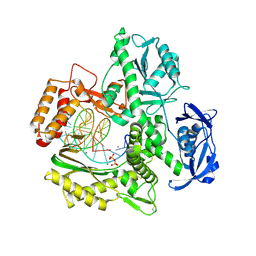 | | Structure of 9N-I DNA polymerase incorporation with dG in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*CP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6ISF
 
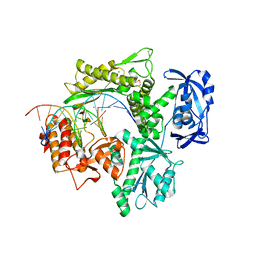 | | Structure of 9N-I DNA polymerase incorporation with dT in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
5XV9
 
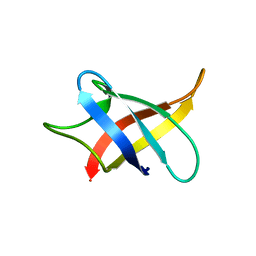 | |
4FJU
 
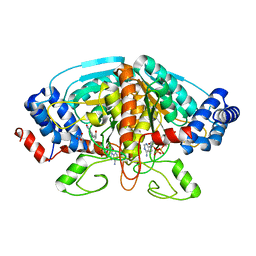 | |
6ISI
 
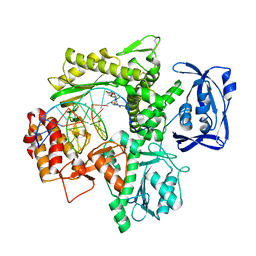 | | Structure of 9N-I DNA polymerase incorporation with 3'-CL in the active site | | Descriptor: | 3-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]propanoic acid, CALCIUM ION, DNA (5'-D(P*AP*CP*GP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
4H8A
 
 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | Authors: | Rhee, S, Shin, I, Kim, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
1BLI
 
 | | BACILLUS LICHENIFORMIS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad.
Structure, 6, 1998
|
|
6ISH
 
 | | Structure of 9N-I DNA polymerase incorporation with 3'-AL in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*(DAL))-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6IS7
 
 | | Structure of 9N-I DNA polymerase incorporation with dA in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*A)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
1IW7
 
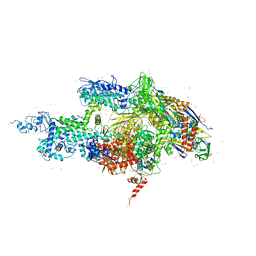 | |
2M0X
 
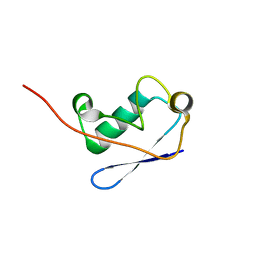 | |
5Z59
 
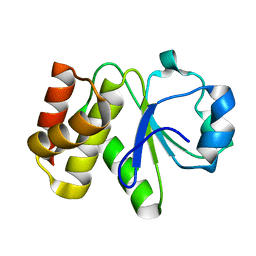 | | Crystal structure of Tk-PTP in the inactive form | | Descriptor: | Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
2NTD
 
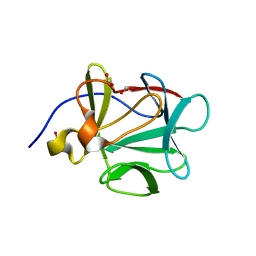 | |
5Z5B
 
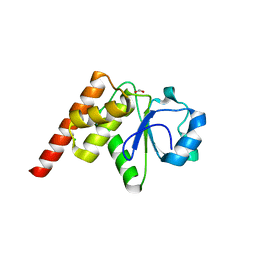 | | Crystal structure of Tk-PTP in the G95A mutant form | | Descriptor: | CHLORIDE ION, FORMIC ACID, Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
5Z5A
 
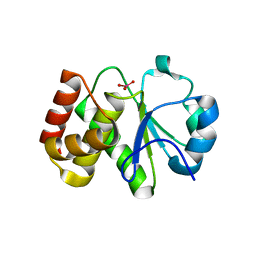 | | Crystal structure of Tk-PTP in the active form | | Descriptor: | Protein-tyrosine phosphatase, VANADATE ION | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
4NQT
 
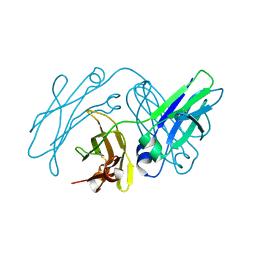 | | anti-parallel Fc-hole(T366S/L368A/Y407V) homodimer | | Descriptor: | Ig gamma-1 chain C region | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-11-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiparallel Conformation of Knob and Hole Aglycosylated Half-Antibody Homodimers Is Mediated by a CH2-CH3 Hydrophobic Interaction.
J.Mol.Biol., 426, 2014
|
|
4NQU
 
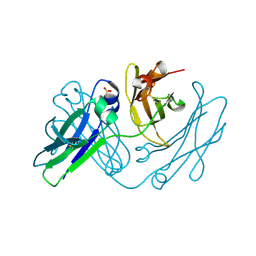 | | anti-parallel Fc-knob (T366W) homodimer | | Descriptor: | Ig gamma-1 chain C region, SULFATE ION | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-11-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antiparallel Conformation of Knob and Hole Aglycosylated Half-Antibody Homodimers Is Mediated by a CH2-CH3 Hydrophobic Interaction.
J.Mol.Biol., 426, 2014
|
|
4NQS
 
 | | Knob-into-hole IgG Fc | | Descriptor: | Ig gamma-1 chain C region, miniZ | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-11-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Antiparallel Conformation of Knob and Hole Aglycosylated Half-Antibody Homodimers Is Mediated by a CH2-CH3 Hydrophobic Interaction.
J.Mol.Biol., 426, 2014
|
|
