1CLU
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1AR7
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR8
 
 | | P1/MAHONEY POLIOVIRUS, MUTANT P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1ASJ
 
 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR9
 
 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
4DHF
 
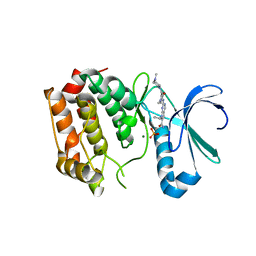 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6VJS
 
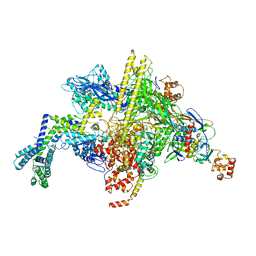 | | Escherichia coli RNA polymerase and ureidothiophene-2-carboxylic acid complex | | Descriptor: | 3-{[benzyl(ethyl)carbamoyl]amino}-5-(4-phenoxyphenyl)thiophene-2-carboxylic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murakami, K.S, Molodtsov, V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus aureus -Based Wound Infection Model in SKH1 Mice.
Acs Infect Dis., 6, 2020
|
|
1AL2
 
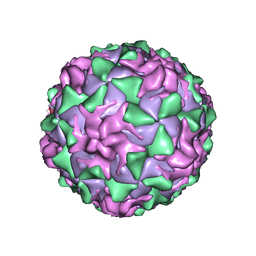 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1XGL
 
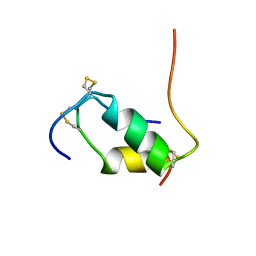 | | HUMAN INSULIN DISULFIDE ISOMER, NMR, 10 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Hua, Q.X, Gozani, S.N, Chance, R.E, Hoffmann, J.A, Frank, B.H, Weiss, M.A. | | Deposit date: | 1996-10-10 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a protein in a kinetic trap.
Nat.Struct.Biol., 2, 1995
|
|
3TH5
 
 | | Crystal structure of wild-type RAC1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
1AR6
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
7U6V
 
 | | Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk | | Descriptor: | C-terminal domain (CTD) from the Ribosomal P-stalk, Shiga toxin 2a subunit A (Stx2A), Shiga toxin 2a subunit B (Stx2B) | | Authors: | Kulczyk, A.W. | | Deposit date: | 2022-03-06 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk reveals residues involved in the binding interaction.
J.Biol.Chem., 299, 2022
|
|
7TTP
 
 | | P450 (OxyA) from kistamicin biosynthesis, mixed heme conformation | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTA
 
 | | P450 (OxyA) from kistamicin biosynthesis, mixed heme conformation, attenuated beam | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTQ
 
 | | P450 (OxyA) from kistamicin biosynthesis, imidazole complex | | Descriptor: | GLYCEROL, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTO
 
 | |
7TTB
 
 | | P450 (OxyA) from kistamicin biosynthesis, Y99F mutant | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801592 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7X6K
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3w | | Descriptor: | 1H-indole-2-carbaldehyde, 3C-like proteinase | | Authors: | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
7X6J
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3af | | Descriptor: | 3C-like proteinase, quinoline-2-carboxylic acid | | Authors: | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
5H4V
 
 | |
8PI3
 
 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
3RYM
 
 | | Structure of Oxidized M98K mutant of Amicyanin | | Descriptor: | Amicyanin, ZINC ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2011-05-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7039 Å) | | Cite: | Replacement of the axial copper ligand methionine with lysine in amicyanin converts it to a zinc-binding protein that no longer binds copper.
J.Inorg.Biochem., 105, 2011
|
|
4JMH
 
 | |
4JMG
 
 | |
1WAR
 
 | | Recombinant Human Purple Acid Phosphatase expressed in Pichia Pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, HUMAN PURPLE ACID PHOSPHATASE, ... | | Authors: | Duff, A.P, Langley, D.B, Han, R, Averill, B.A, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-10-28 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structures of Recombinant Human Purple Acid Phosphatase with and without an Inhibitory Conformation of the Repression Loop.
J.Mol.Biol., 351, 2005
|
|
