3M4J
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5E
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[N-(6-chloro-5-formyl-2-methylthiopyrimidin-4-yl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-({[6-chloro-5-formyl-2-(methylsulfanyl)pyrimidin-4-yl]amino}methyl)benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
6I47
 
 | | Structure of P. aeruginosa LpxC with compound 10: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-1-oxoisoindolin-2-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, (2~{S})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
3C96
 
 | | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR240 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa.
To be Published
|
|
6I49
 
 | | Structure of P. aeruginosa LpxC with compound 17a: (2R)-N-Hydroxy-2-methyl-2-(methylsulfonyl)-4(6((4(morpholinomethyl)phenyl)ethynyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)yl)butanamide | | Descriptor: | (2~{R})-2-methyl-2-methylsulfonyl-4-[6-[2-[4-(morpholin-4-ylmethyl)phenyl]ethynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
3C5R
 
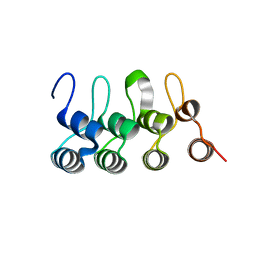 | |
1JAZ
 
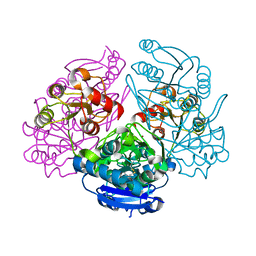 | |
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | Descriptor: | De Novo design cysteine esterase FR29 | | Authors: | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3BRH
 
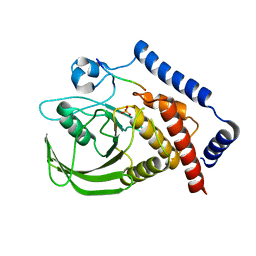 | |
1K57
 
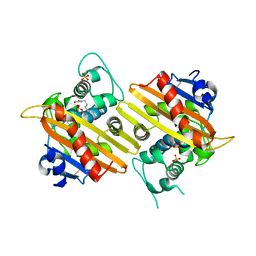 | | OXA 10 class D beta-lactamase at pH 6.0 | | Descriptor: | BETA LACTAMASE OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K56
 
 | | OXA 10 class D beta-lactamase at pH 6.5 | | Descriptor: | OXA10 beta-lactamase, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3NHD
 
 | | GYVLGS segment 127-132 from human prion with V129 | | Descriptor: | ACETIC ACID, Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
3CLR
 
 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
4HB5
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR267. | | Descriptor: | Engineered Protein | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
3CW9
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase in the Thioester-forming Conformation, bound to 4-chlorophenacyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-Chlorophenacyl-coenzyme A, 4-chlorobenzoyl CoA ligase, ... | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NVH
 
 | |
1IGB
 
 | | AEROMONAS PROTEOLYTICA AMINOPEPTIDASE COMPLEXED WITH THE INHIBITOR PARA-IODO-D-PHENYLALANINE HYDROXAMATE | | Descriptor: | AMINOPEPTIDASE, PARA-IODO-D-PHENYLALANINE HYDROXAMIC ACID, ZINC ION | | Authors: | Chevrier, B, D'Orchymont, H, Schalk, C, Tarnus, C, Moras, D. | | Deposit date: | 1996-02-27 | | Release date: | 1996-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit.
Eur.J.Biochem., 237, 1996
|
|
3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
3CLU
 
 | | Crystal structure of the R236K mutant from Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
1KRW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1HTV
 
 | |
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
