2IEO
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Protease, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
2IJD
 
 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
1D4V
 
 | | Crystal structure of trail-DR5 complex | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Mongkolsapaya, J, Grimes, J.M, Stuart, D.I, Jones, E.Y, Screaton, G.R. | | Deposit date: | 1999-10-06 | | Release date: | 1999-11-01 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation
Nat.Struct.Biol., 6, 1999
|
|
2IM5
 
 | | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas Gingivalis | | Descriptor: | Nicotinate phosphoribosyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-03 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas gingivalis
To be Published
|
|
1RSD
 
 | | DHNA complex with 3-(5-Amino-7-hydroxy-[1,2,3]triazolo[4,5-d]pyrimidin-2-yl)-N-[2-(2-hydroxymethyl-phenylsulfanyl)-benzyl]-benzamide | | Descriptor: | 3-(5-AMINO-7-HYDROXY-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-2-YL)-N-[2-(2-(HYDROXYMETHYL-PHENYLSULFANYL)-BENZYL]-BENZAMIDE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-09 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
2IJF
 
 | |
1R6L
 
 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ribonuclease PH, SULFATE ION | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
2IM9
 
 | | Crystal structure of protein LPG0564 from Legionella pneumophila str. Philadelphia 1, Pfam DUF1460 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of hypothetical protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
TO BE PUBLISHED
|
|
1R70
 
 | | Model of human IgA2 determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Human IgA2(m1) Heavy Chain, Human IgA2(m1) Light Chain | | Authors: | Furtado, P.B, Whitty, P.W, Robertson, A, Eaton, J.T, Almogren, A, Kerr, M.A, Woof, J.M, Perkins, S.J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | Solution Structure Determination of Monomeric Human IgA2 by X-ray and Neutron Scattering, Analytical Ultracentrifugation and Constrained Modelling: A Comparison with Monomeric Human IgA1.
J.Mol.Biol., 338, 2004
|
|
2IS7
 
 | | Crystal Structure of Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Milne, A, Brownlee, J.M. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
1R44
 
 | | Crystal Structure of VanX | | Descriptor: | D-alanyl-D-alanine dipeptidase, ZINC ION | | Authors: | Pratt, S.D, Katz, L, Severin, J.M, Holzman, T, Park, C.H. | | Deposit date: | 2003-10-03 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of VanX Reveals a Novel Amino-Dipeptidase Involved in Mediating Transposon-Based Vancomycin Resistance
Mol.Cell, 2, 1998
|
|
1R4N
 
 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like protein NEDD8, ZINC ION, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
1REJ
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1A16
 
 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AG7
 
 | | CONOTOXIN GS, NMR, 20 STRUCTURES | | Descriptor: | CONOTOXIN GS | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1997-04-03 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel antagonist conotoxin GS: a new molecular caliper for probing sodium channel geometry.
Structure, 5, 1997
|
|
2K5V
 
 | | SOLUTION NMR STRUCTURE OF the second OB-fold domain of replication protein A from Methanococcus maripaludis. NORTHEAST STRUCTURAL GENOMICS TARGET MrR110B. | | Descriptor: | Replication protein A | | Authors: | Aramini, J.M, Maglaqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.VT, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF the second OB-fold domain of replication protein A from Methanococcus maripaludis. NORTHEAST STRUCTURAL GENOMICS TARGET MrR110B.
To be Published
|
|
2KA6
 
 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
1SX1
 
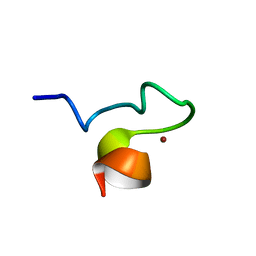 | | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase | | Descriptor: | SecA, ZINC ION | | Authors: | Dempsey, B.R, Wrona, M, Moulin, J.M, Gloor, G.B, Jalilehvand, F, Lajoie, G, Shaw, G.S, Shilton, B.H. | | Deposit date: | 2004-03-30 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase.
Biochemistry, 43, 2004
|
|
1T1S
 
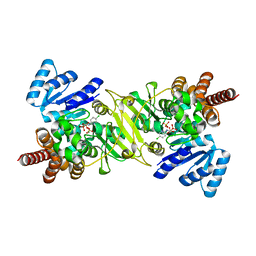 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
2KT1
 
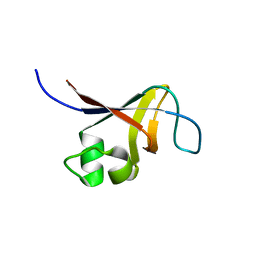 | | Solution NMR Structure of the SH3 Domain from the p85beta subunit of Phosphatidylinositol 3-kinase from H.sapiens, Northeast Structural Genomics Consortium Target HR5531E | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Aramini, J.M, Ma, L, Schauder, C, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-15 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the SH3 Domain from the p85beta subunit of Phosphatidylinositol 3-kinase from H.sapiens, Northeast Structural Genomics Consortium Target HR5531E
To be Published
|
|
1D3Y
 
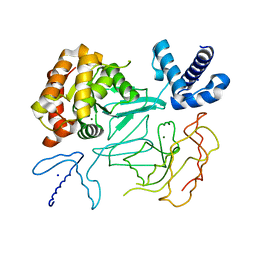 | | STRUCTURE OF THE DNA TOPOISOMERASE VI A SUBUNIT | | Descriptor: | DNA TOPOISOMERASE VI A SUBUNIT, MAGNESIUM ION, SODIUM ION | | Authors: | Nichols, M.D, DeAngelis, K.A, Keck, J.L, Berger, J.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11.
EMBO J., 18, 1999
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I5G
 
 | | Crystal strcuture of amidohydrolase from Pseudomonas aeruginosa | | Descriptor: | amidohydrolase | | Authors: | Min, T, Sauder, J.M, Wasserman, S.R, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of amidohydrolase from Pseudomonas aeruginosa
To be Published
|
|
2I52
 
 | | Crystal structure of protein PTO0218 from Picrophilus torridus, Pfam DUF372 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramagopal, U.A, Gilmore, J, Toro, R, Bain, K.T, McKenzie, C, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-23 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of hypothetical protein PTO0218 from Picrophilus torridus
To be Published
|
|
1D3B
 
 | | CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B), ... | | Authors: | Kambach, C, Walke, S, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
