3ZPH
 
 | |
1DAG
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID AND 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1BYI
 
 | | STRUCTURE OF APO-DETHIOBIOTIN SYNTHASE AT 0.97 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHASE | | Authors: | Sandalova, T, Schneider, G, Kaeck, H, Lindqvist, Y. | | Deposit date: | 1998-10-15 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of dethiobiotin synthetase at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2OE7
 
 | | High-Pressure T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
1VSM
 
 | |
8QYQ
 
 | | Beta-cardiac myosin S1 fragment in the pre-powerstroke state complexed to Mavacamten | | Descriptor: | 6-[[(1~{S})-1-phenylethyl]amino]-3-propan-2-yl-1~{H}-pyrimidine-2,4-dione, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
8QYP
 
 | | Beta-cardiac myosin motor domain in the pre-powerstroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-7, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
8QYU
 
 | | Beta-cardiac myosin S1 fragment in the pre-powerstroke state complexed to Omecamtiv mecarbil | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
8QYR
 
 | | Beta-cardiac myosin motor domain in the pre-powerstroke state complexed to Mavacamten | | Descriptor: | 1,2-ETHANEDIOL, 6-[[(1~{S})-1-phenylethyl]amino]-3-propan-2-yl-1~{H}-pyrimidine-2,4-dione, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
3PFW
 
 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD, a binary form | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
7JYZ
 
 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | Descriptor: | Bromodomain-containing protein 3, Integrase peptide | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JMY
 
 | |
7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
2ITG
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
1VSK
 
 | |
1VSL
 
 | | ASV INTEGRASE CORE DOMAIN D64N MUTATION WITH ZINC CATION | | Descriptor: | PROTEIN (INTEGRASE), ZINC ION | | Authors: | Lubkowski, J, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inactivating mutations and pH-dependent activity of avian sarcoma virus integrase.
J.Biol.Chem., 273, 1998
|
|
8PEE
 
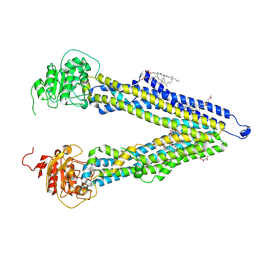 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
3M1S
 
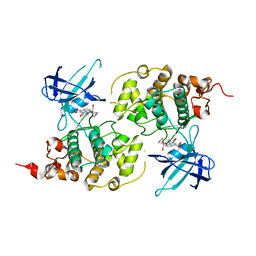 | | Structure of Ruthenium Half-Sandwich Complex Bound to Glycogen Synthase Kinase 3 | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium pyridocarbazole | | Authors: | Atilla-Gokcumen, G.E, Di Costanzo, L, Zimmermann, G, Meggers, E. | | Deposit date: | 2010-03-05 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.134 Å) | | Cite: | Structure of anticancer ruthenium half-sandwich complex bound to glycogen synthase kinase 3beta
J.Biol.Inorg.Chem., 16, 2011
|
|
1BQM
 
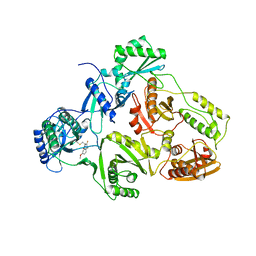 | | HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQN
 
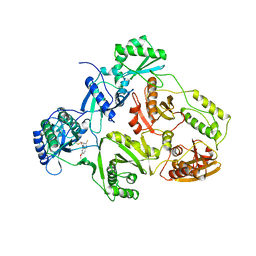 | | TYR 188 LEU HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
3RUG
 
 | | Crystal structure of Valpha10-Vbeta8.1 NKT TCR in complex with CD1d-alphaglucosylceramide (C20:2) | | Descriptor: | (11E,14E)-N-[(2S,3S,4R)-1-(alpha-D-glucopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A semi-invariant V(alpha)10(+) T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
Nat.Immunol., 12, 2011
|
|
2OE9
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
2IB9
 
 | |
2IBY
 
 | |
