1R5O
 
 | | crystal structure analysis of sup35 complexed with GMPPNP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-11 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
6QJ5
 
 | | X-ray structure of PPARgamma LBD with the ligand NV1380 | | Descriptor: | (2~{S})-3-methyl-2-[(4-octoxyphenyl)carbonylamino]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D. | | Deposit date: | 2019-01-22 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor gamma Binding Properties and Biological Activities.
J.Med.Chem., 63, 2020
|
|
3E1Y
 
 | | Crystal structure of human eRF1/eRF3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
3E20
 
 | | Crystal structure of S.pombe eRF1/eRF3 complex | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
1D5M
 
 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDE AND SEB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Swain, A.L, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-07 | | Release date: | 2000-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
1D5X
 
 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH DIPEPTIDE MIMETIC AND SEB | | Descriptor: | DIPEPTIDE MIMETIC INHIBITOR, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
8GZC
 
 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
8JS9
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Botulinum neurotoxin, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLF
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Botulinum neurotoxin, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLE
 
 | |
8JS8
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLG
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Botulinum neurotoxin, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLC
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLH
 
 | | Cryo-EM structure of SV2A dimer in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8K77
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc and brivaracetam | | Descriptor: | (2S)-2-[(4R)-2-oxidanylidene-4-propyl-pyrrolidin-1-yl]butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-07-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JLI
 
 | | Cryo-EM structure of SV2A dimer in complex levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, Synaptic vesicle glycoprotein 2A | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
5Y50
 
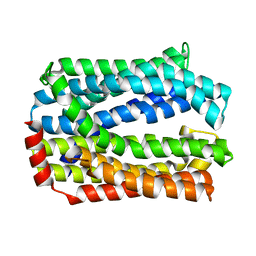 | | Crystal structure of eukaryotic MATE transporter AtDTX14 | | Descriptor: | Protein DETOXIFICATION 14 | | Authors: | Miyauchi, H, Kusakizako, T, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for xenobiotic extrusion by eukaryotic MATE transporter
Nat Commun, 8, 2017
|
|
6SZS
 
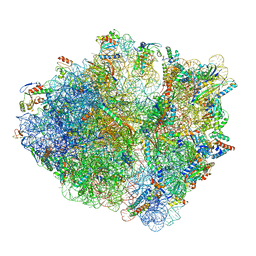 | | Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Muller, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis.
Nat Commun, 10, 2019
|
|
3NGO
 
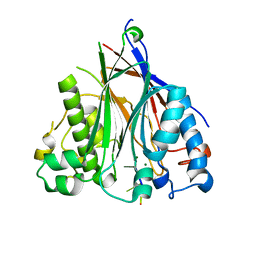 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
5XDO
 
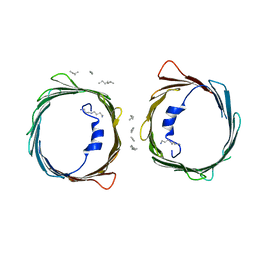 | | Crystal structure of human voltage-dependent anion channel 1 (hVDAC1) in C222 space group | | Descriptor: | HEXANE, N-OCTANE, PENTANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structural characterization reveals novel oligomeric interactions of human voltage-dependent anion channel 1
Protein Sci., 26, 2017
|
|
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
5B2G
 
 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
5XDN
 
 | | Crystal structure of human voltage-dependent anion channel 1 (hVDAC1) in P22121 space group | | Descriptor: | DECANE, DODECANE, HEXANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structural characterization reveals novel oligomeric interactions of human voltage-dependent anion channel 1
Protein Sci., 26, 2017
|
|
8JH8
 
 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Feruloyl esterase, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
