6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6TOH
 
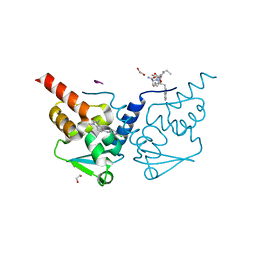 | | Crystal structure of human BCL6 BTB domain in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1,3-dimethyl-2-oxidanylidene-benzimidazol-5-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Shetty, K, Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6TOI
 
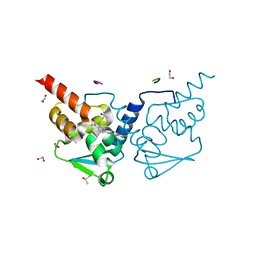 | | Crystal structure of human BCL6 BTB domain in complex with compound 11f | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-3-[(3~{R})-3-oxidanylbutyl]-2-oxidanylidene-benzimidazol-5-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6TOO
 
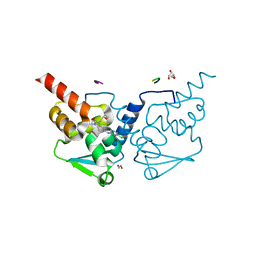 | | Crystal structure of human BCL6 BTB domain in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-3-[(2~{S})-2-oxidanylbutyl]-2-oxidanylidene-benzimidazol-5-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6CKQ
 
 | |
6SUC
 
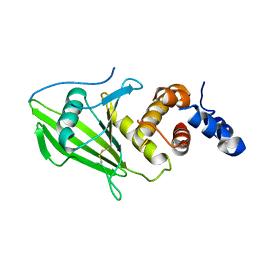 | | Human PTPRU D1 domain, oxidised form | | Descriptor: | Receptor-type tyrosine-protein phosphatase U | | Authors: | Hay, I.M, Fearnley, G.W, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The receptor PTPRU is a redox sensitive pseudophosphatase.
Nat Commun, 11, 2020
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6SUB
 
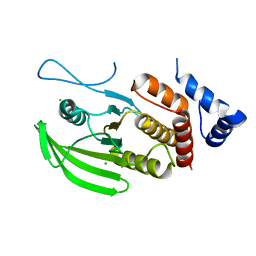 | | Human PTPRU D1 domain, reduced form | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase U | | Authors: | Hay, I.M, Fearnley, G.W, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The receptor PTPRU is a redox sensitive pseudophosphatase.
Nat Commun, 11, 2020
|
|
6UO9
 
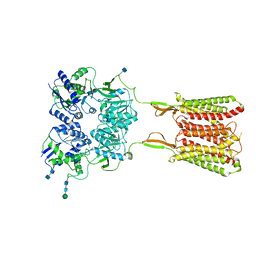 | | Human metabotropic GABA(B) receptor bound to agonist SKF97541 in its intermediate state 2 | | Descriptor: | (R)-(3-aminopropyl)methylphosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaye, H, Han, G.W, Gati, C, Cherezov, V. | | Deposit date: | 2019-10-14 | | Release date: | 2020-06-10 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of a metabotropic GABA receptor.
Nature, 584, 2020
|
|
6UOA
 
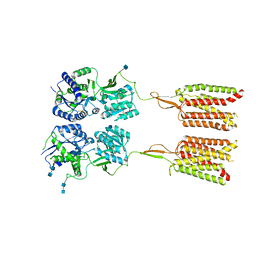 | | Human metabotropic GABA(B) receptor in its intermediate state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Shaye, H, Han, G.W, Gati, C, Cherezov, V. | | Deposit date: | 2019-10-14 | | Release date: | 2020-06-10 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural basis of the activation of a metabotropic GABA receptor.
Nature, 584, 2020
|
|
6UHW
 
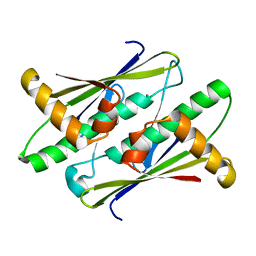 | |
6U6G
 
 | |
6UO8
 
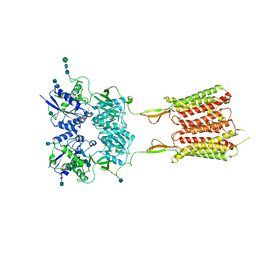 | | Human metabotropic GABA(B) receptor bound to agonist SKF97541 and positive allosteric modulator GS39783 | | Descriptor: | (R)-(3-aminopropyl)methylphosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaye, H, Han, G.W, Gati, C, Cherezov, V. | | Deposit date: | 2019-10-14 | | Release date: | 2020-06-10 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis of the activation of a metabotropic GABA receptor.
Nature, 584, 2020
|
|
6ECD
 
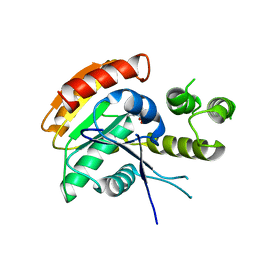 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a tetradepsipeptide | | Descriptor: | Vlm2, tetradepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
6ECF
 
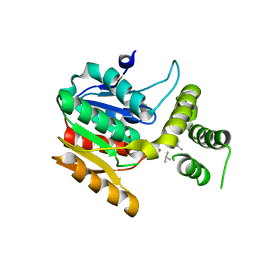 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a dodecadepsipeptide, space group P1 | | Descriptor: | Vlm2, dodecadepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
6BQH
 
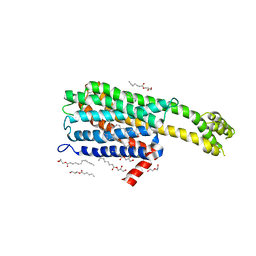 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
2GIC
 
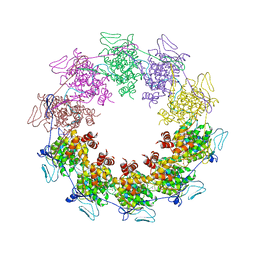 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
2IWE
 
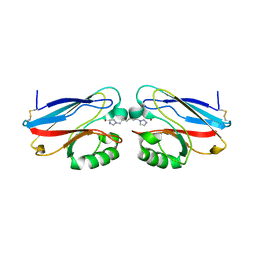 | | Structure of a cavity mutant (H117G) of Pseudomonas aeruginosa azurin | | Descriptor: | 1,1'-HEXANE-1,6-DIYLBIS(1H-IMIDAZOLE), AZURIN, ZINC ION | | Authors: | De Jongh, T.E, Van Roon, A.M.M, Prudencio, M, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Click-Chemistry with an Active Site Variant of Azurin
Eur.J.Inorg.Chem., 2006, 2006
|
|
2F3L
 
 | |
2F05
 
 | |
2FWU
 
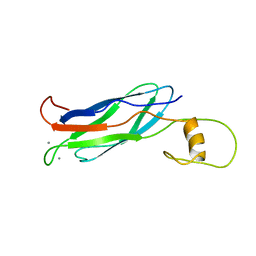 | |
2G0Y
 
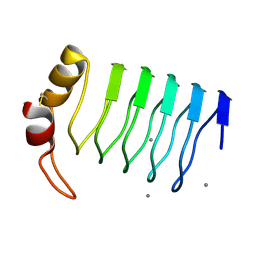 | | Crystal Structure of a Lumenal Pentapeptide Repeat Protein from Cyanothece sp 51142 at 2.3 Angstrom Resolution. Tetragonal Crystal Form | | Descriptor: | CALCIUM ION, pentapeptide repeat protein | | Authors: | Kennedy, M.A, Ni, S, Buchko, G.W, Robinson, H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of two potentially universal turn motifs that shape the repeated five-residues fold - Crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142
Protein Sci., 15, 2006
|
|
2GM7
 
 | | TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Ten A Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
2FWS
 
 | |
