7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5TZD
 
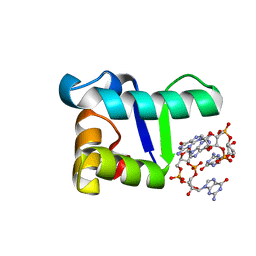 | | Structure of the WT S. venezulae BldD-(CTD-c-di-GMP)2 assembly intermediate | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7OMU
 
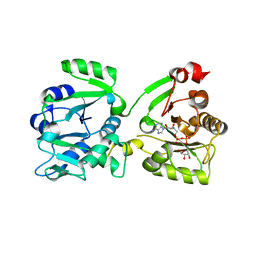 | | Thermosipho africanus DarTG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Macro domain-containing protein | | Authors: | Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
5K58
 
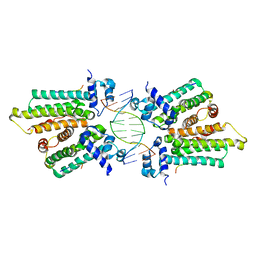 | |
5K5O
 
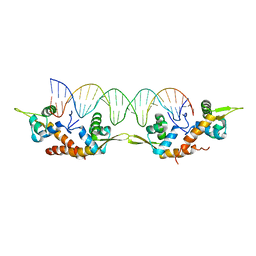 | | Structure of AspA-26mer DNA complex | | Descriptor: | AspA, DNA (26-MER) | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5A
 
 | | Structure of the pNOB8-like ParB N-domain | | Descriptor: | ParB domain protein nuclease | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.825 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5D
 
 | | Structure of the C2221 form of Pnob8-like ParB-N domain | | Descriptor: | CITRIC ACID, ParB domain protein nuclease | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5R
 
 | | AspA-32mer DNA,crystal form 2 | | Descriptor: | AspA, DNA (32-MER), PHOSPHATE ION | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5Q
 
 | |
5KHD
 
 | | Structure of 1.75 A BldD C-domain-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7501 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell, 158, 2014
|
|
5K5Z
 
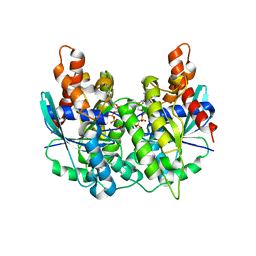 | | Structure of pnob8 ParA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.369 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5KBJ
 
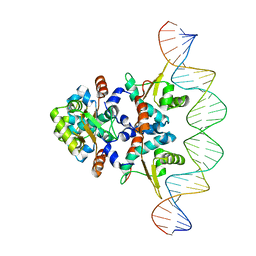 | | Structure of Rep-DNA complex | | Descriptor: | DNA (32-MER), Replication initiator A, N-terminal | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
5J6E
 
 | |
7ZAX
 
 | |
7QS6
 
 | |
7ZED
 
 | |
8BSS
 
 | | Solution Structure of thanatin-like derivative 5 in complex with E. coli LptA mutant Q62L | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin-like derivative | | Authors: | Oi, K.K, Jurt, S, Moehle, K, Zerbe, O. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Peptidomimetic antibiotics disrupt the lipopolysaccharide transport bridge of drug-resistant Enterobacteriaceae.
Sci Adv, 9, 2023
|
|
5V53
 
 | |
6Q9C
 
 | | Crystal structure of Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF bound to NADH under anaerobic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9G
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129D and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9J
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129S and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9K
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF S96M bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
