4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
1R0U
 
 | | Crystal structure of ywiB protein from Bacillus subtilis | | Descriptor: | GLYCEROL, protein ywiB | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of ywiB protein from Bacillus subtilis
to be published
|
|
1T9K
 
 | | X-ray crystal structure of aIF-2B alpha subunit-related translation initiation factor [Thermotoga maritima] | | Descriptor: | CHLORIDE ION, Probable methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of aIF-2B translation initiation factor from Thermotoga maritima
To be Published
|
|
4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
2BB3
 
 | | Crystal Structure of Cobalamin Biosynthesis Precorrin-6Y Methylase (cbiE) from Archaeoglobus fulgidus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, cobalamin biosynthesis precorrin-6Y methylase (cbiE) | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Cobalamin Biosynthesis Precorrin-6Y Methylase (cbiE) from Archaeoglobus fulgidus
To be Published
|
|
1PF5
 
 | | Structural Genomics, Protein YJGH | | Descriptor: | Hypothetical protein yjgH, MERCURY (II) ION | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Xu, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5A crystal structure of protein YJGH from E. Coli
To be Published
|
|
1PBJ
 
 | | CBS domain protein | | Descriptor: | MAGNESIUM ION, hypothetical protein | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-14 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a hypothetical protein from M. thermautotrophicus reveals a
novel fold and a pseudo 2-fold axis of symmetry
TO BE PUBLISHED
|
|
1PBT
 
 | | The crystal structure of TM1154, oxidoreductase, sol/devB family from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-phosphogluconolactonase, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure analysis of TM1154, oxidoreductase from Thermotoga maritima
To be Published
|
|
1KS2
 
 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
2HS5
 
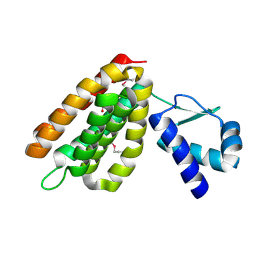 | | Structural Genomics, the crystal structure of a putative transcriptional regulator GntR from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, putative transcriptional regulator GntR | | Authors: | Tan, K, Skarina, T, Onopriyenko, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-21 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a putative transcriptional regulator GntR from Rhodococcus sp. RHA1
To be Published
|
|
1TP6
 
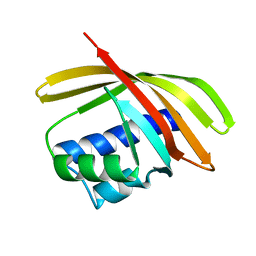 | | 1.5 A Crystal Structure of a NTF-2 Like Protein of Unknown Function PA1314 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1314 | | Authors: | Zhang, R, Xu, L.X, savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa
To be Published
|
|
1TU1
 
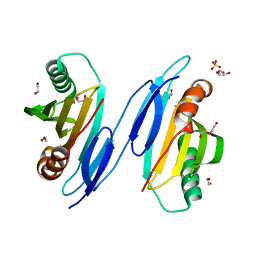 | | Crystal Structure of Protein of Unknown Function PA94 from Pseudomonas aeruginosa, Putative Regulator | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA94 from Pseudomonas aeruginosa
To be Published
|
|
1TUA
 
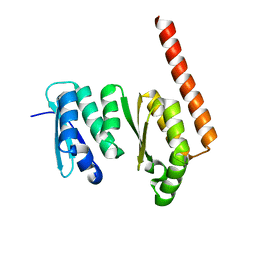 | | 1.5 A Crystal Structure of a Protein of Unknown Function APE0754 from Aeropyrum pernix | | Descriptor: | Hypothetical protein APE0754 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix
To be Published
|
|
2LEZ
 
 | | Solution NMR structure of N-terminal domain of Salmonella effector protein PipB2. Northeast structural genomics consortium (NESG) target stt318a | | Descriptor: | Secreted effector protein pipB2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Daniels, C, Savchenko, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of Salmonella effector protein PipB2
To be Published
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2NP3
 
 | | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3. | | Descriptor: | Putative TetR-family regulator | | Authors: | Koclega, K.D, Xu, X, Chruszcz, M, Gu, J, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3.
To be Published
|
|
2NP5
 
 | | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1.
TO BE PUBLISHED
|
|
2O30
 
 | | Nuclear movement protein from E. cuniculi GB-M1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NUCLEAR MOVEMENT PROTEIN | | Authors: | Binkowski, T.A, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Nuclear movement protein from E. cuniculi GB-M1
TO BE PUBLISHED
|
|
2O38
 
 | | Putative XRE Family Transcriptional Regulator | | Descriptor: | ACETIC ACID, Hypothetical protein | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The Crystal Structure of Putative XRE Family Transcriptional Regulator
To be Published
|
|
2OJH
 
 | | The structure of putative TolB from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, Uncharacterized protein Atu1656/AGR_C_3050 | | Authors: | Cuff, M.E, Xu, X, Gu, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of putative TolB from Agrobacterium tumefaciens
TO BE PUBLISHED
|
|
2OR0
 
 | | Structural Genomics, the crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, Hydroxylase | | Authors: | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1
To be Published
|
|
2OMO
 
 | | Putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea | | Descriptor: | DUF176 | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-22 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea.
To be Published
|
|
2OQG
 
 | | ArsR-like Transcriptional Regulator from Rhodococcus sp. RHA1 | | Descriptor: | ACETIC ACID, Possible transcriptional regulator, ArsR family protein | | Authors: | Kim, Y, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the ArsR-like Transcriptional Regulator from Rhodococcus sp. RHA1
To be Published
|
|
3TTG
 
 | | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, Putative aminomethyltransferase | | Authors: | Michalska, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum
TO BE PUBLISHED
|
|
3HE1
 
 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
