7YFY
 
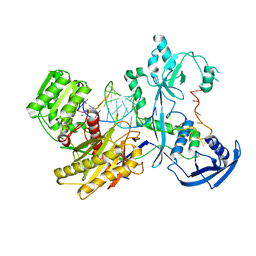 | | Cryo-EM structure of the Mili-piRNA- target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (5'-R(P*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
9D8U
 
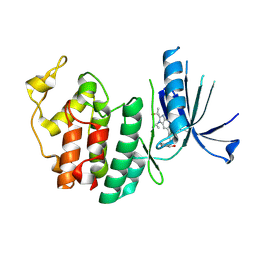 | | Crystal structure of CDK6 in complex with atirmociclib | | Descriptor: | Atirmociclib, Cyclin-dependent kinase 6 | | Authors: | Johnson, E, Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2024-08-20 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDK4 selective inhibition improves preclinical anti-tumor efficacy and safety.
Cancer Cell, 43, 2025
|
|
3M3O
 
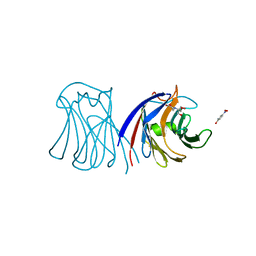 | | Crystal Structure of Agrocybe aegerita lectin AAL mutant R85A complexed with p-Nitrophenyl TF disaccharide | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-tumor lectin, P-NITROPHENOL, ... | | Authors: | Feng, L, Li, D, Wang, D. | | Deposit date: | 2010-03-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen
Faseb J., 24, 2010
|
|
8WE1
 
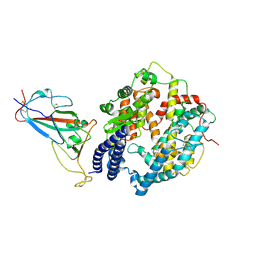 | | SARS-CoV-2 Omicron BF.7 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8WDZ
 
 | | SARS-CoV-2 Omicron BQ.1 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8WE4
 
 | | SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-17 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8WDS
 
 | | Crystal structure of BF.7 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8WDR
 
 | | Crystal structure of BQ.1.1 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8WE0
 
 | | SARS-CoV-2 Omicron XBB RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8WDY
 
 | | SARS-CoV-2 Omicron BQ.1.1 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8XJV
 
 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
3M3C
 
 | | Crystal Structure of Agrocybe aegerita lectin AAL complexed with p-Nitrophenyl TF disaccharide | | Descriptor: | Anti-tumor lectin, P-NITROPHENOL, SULFATE ION, ... | | Authors: | Feng, L, Li, D, Wang, D. | | Deposit date: | 2010-03-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen
Faseb J., 24, 2010
|
|
3M3E
 
 | |
1SVM
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
3M3Q
 
 | |
1SVO
 
 | | Structure of SV40 large T antigen helicase domain | | Descriptor: | ZINC ION, large T antigen | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
4HJX
 
 | | Crystal structure of F2YRS complexed with F2Y | | Descriptor: | 3,5-difluoro-L-tyrosine, Tyrosine-tRNA ligase | | Authors: | Wang, J, Tian, C, Gong, W, Li, F, Shi, P, Li, J, Ding, W. | | Deposit date: | 2012-10-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A genetically encoded 19F NMR probe for tyrosine phosphorylation.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1SVL
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
3U28
 
 | | Crystal structure of a Cbf5-Nop10-Gar1 complex from Saccharomyces cerevisiae | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 3, H/ACA ribonucleoprotein complex subunit 4 | | Authors: | Ye, K, Li, S. | | Deposit date: | 2011-10-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase
Genes Dev., 25, 2011
|
|
6UCA
 
 | | Crystal structure of human ZCCHC4 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, rRNA N6-adenosine-methyltransferase ZCCHC4 | | Authors: | Lu, J.W, Ren, W.D, Huang, M.J, Gao, L, Li, D.X, Wang, G.G, Song, J. | | Deposit date: | 2019-09-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA.
Nat Commun, 10, 2019
|
|
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
6UUM
 
 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
8DTK
 
 | |
6UUL
 
 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
