6BFB
 
 | | Crystal structure of a F. nucleatum FMN riboswitch bound to WG-3 | | Descriptor: | 5-[(3S,4S)-3-(dimethylamino)-4-hydroxypyrrolidin-1-yl]-6-fluoro-4-methyl-8-oxo-3,4-dihydro-8H-1-thia-4,9b-diazacyclopenta[cd]phenalene-9-carboxylic acid, MAGNESIUM ION, RNA (54-MER), ... | | Authors: | Rizvi, N.F, Fischmann, T.O. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry.
ACS Chem. Biol., 13, 2018
|
|
6VK3
 
 | | CryoEM structure of Hrd3/Yos9 complex | | Descriptor: | Hrd3, Protein OS-9 homolog | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VJZ
 
 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 complex of the expected topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VK0
 
 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 of the flipped topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VJY
 
 | | Cryo-EM structure of Hrd1/Hrd3 monomer | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
3NDB
 
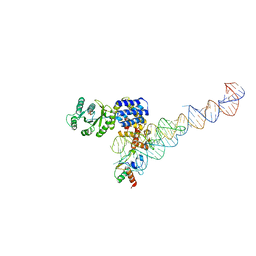 | | Crystal structure of a signal sequence bound to the signal recognition particle | | Descriptor: | PHOSPHATE ION, SRP RNA, Signal recognition 54 kDa protein, ... | | Authors: | Hainzl, T, Huang, S, Sauer-Eriksson, E. | | Deposit date: | 2010-06-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of signal-sequence recognition by the signal recognition particle.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6VK1
 
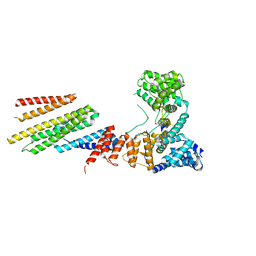 | | CryoEM structure of Hrd1/Hrd3 part from Hrd1-Usa1/Der1/Hrd3 complex | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
3PFW
 
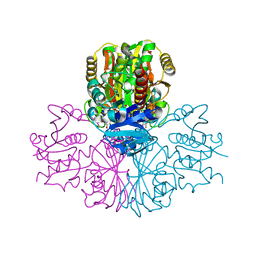 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD, a binary form | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
1DAG
 
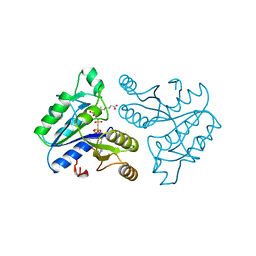 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID AND 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAH
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7,8-DIAMINO-NONANOIC ACID, 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE, AND MANGANESE | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, MANGANESE (II) ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAI
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
7YMV
 
 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.74 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMW
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMX
 
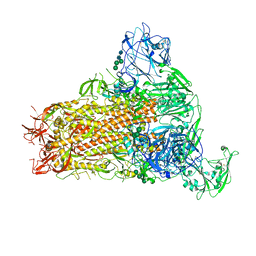 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMY
 
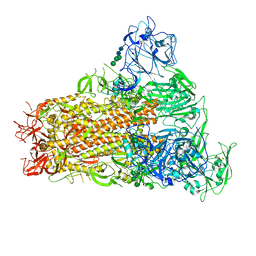 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMZ
 
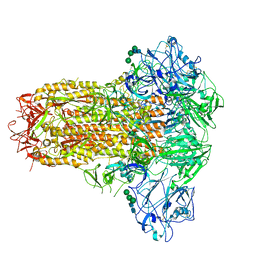 | | Cryo-EM structure of MERS-CoV spike protein, intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMT
 
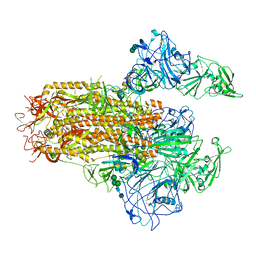 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.55 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YN0
 
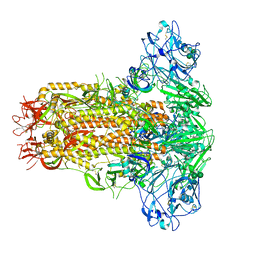 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | Descriptor: | Bromodomain-containing protein 3, Integrase peptide | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JMY
 
 | |
7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
1DAF
 
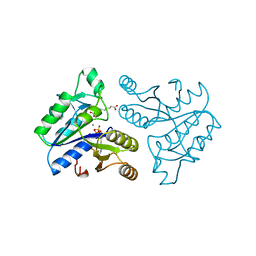 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADP, AND CALCIUM | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAE
 
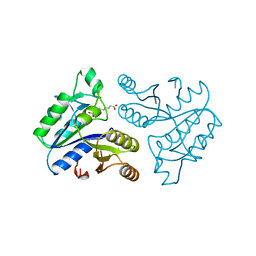 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 3-(1-AMINOETHYL) NONANEDIOIC ACID | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
7ZKA
 
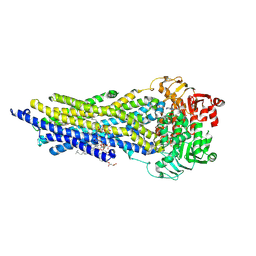 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK5
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
