4L1W
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
4L1X
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 V54L Mutant in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
4JM7
 
 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
9BXA
 
 | |
5N3U
 
 | | The structure of the complex of CpcE and CpcF of phycocyanin lyase from Nostoc sp. PCC7120 | | Descriptor: | Phycocyanobilin lyase subunit alpha, Phycocyanobilin lyase subunit beta | | Authors: | Hoeppner, A, Zhao, C, Xu, Q.-Z, Gaertner, W, Scheer, H, Zhao, K.-H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3RHZ
 
 | |
1ECP
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-13 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.
Structure, 5, 1997
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1PBN
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-10 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
4XXI
 
 | | Crystal structure of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | Descriptor: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | Authors: | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | Deposit date: | 2015-01-30 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1QHT
 
 | |
4RXJ
 
 | | crystal structure of WHSC1L1-PWWP2 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
8I71
 
 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
7MWI
 
 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
2KFT
 
 | |
7CH6
 
 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH8
 
 | | Cryo-EM structure of P.aeruginosa MlaFEBD with ADP-V | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH9
 
 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CHA
 
 | | Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH7
 
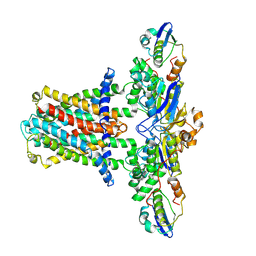 | | Cryo-EM structure of E.coli MlaFEB | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, Phospholipid ABC transporter ATP-binding protein MlaF | | Authors: | Zhou, C, Shi, H, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
227D
 
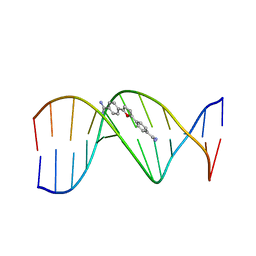 | | A CRYSTALLOGRAPHIC AND SPECTROSCOPIC STUDY OF THE COMPLEX BETWEEN D(CGCGAATTCGCG)2 AND 2,5-BIS(4-GUANYLPHENYL)FURAN, AN ANALOGUE OF BERENIL. STRUCTURAL ORIGINS OF ENHANCED DNA-BINDING AFFINITY | | Descriptor: | 2,5-BIS(4-GUANYLPHENYL)FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Laughton, C.A, Tanious, F, Nunn, C.M, Boykin, D.W, Wilson, W.D, Neidle, S. | | Deposit date: | 1995-08-08 | | Release date: | 1995-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity.
Biochemistry, 35, 1996
|
|
5TCZ
 
 | |
6V3F
 
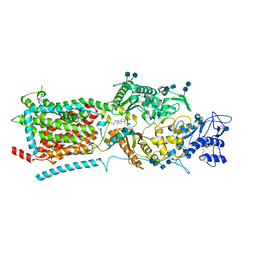 | | Structure of NPC1-like intracellular cholesterol transporter 1 (NPC1L1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z. | | Deposit date: | 2019-11-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition
Sci Adv, 6, 2020
|
|
