3QJY
 
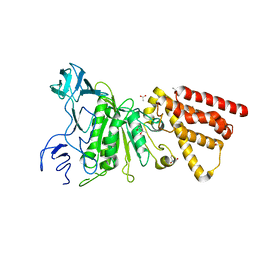 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
2K1F
 
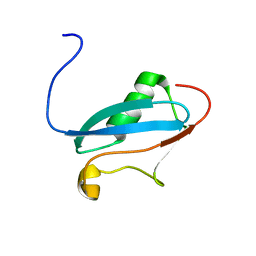 | | SUMO-3 from Drosophila melanogaster (dsmt3) | | Descriptor: | CG4494-PA | | Authors: | Kumar, D, Misra, J.R, Misra, A.K, Chugh, J, Sharma, S, Hosur, R.V. | | Deposit date: | 2008-03-03 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR-derived solution structure of SUMO from Drosophila melanogaster (dSmt3).
Proteins, 75, 2009
|
|
5D7V
 
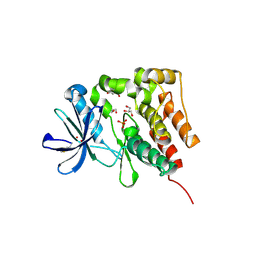 | | Crystal structure of PTK6 kinase domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the kinase domain of human protein tyrosine kinase 6 (PTK6) at 2.33 angstrom resolution
Biochem.Biophys.Res.Commun., 478, 2016
|
|
2MI1
 
 | |
3SSA
 
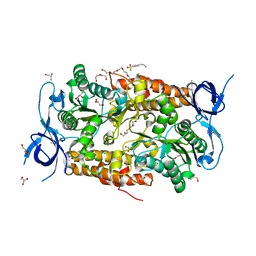 | | Crystal structure of subunit B mutant N157T of the A1AO ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sundararaman, L, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-07-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
8ICW
 
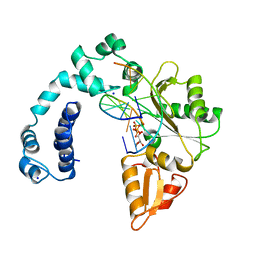 | |
3TGW
 
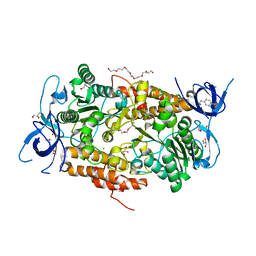 | | Crystal structure of subunit B mutant H156A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
8ICY
 
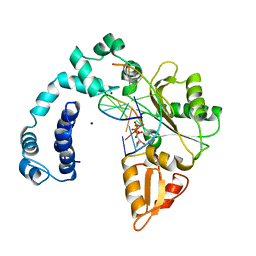 | |
8ICT
 
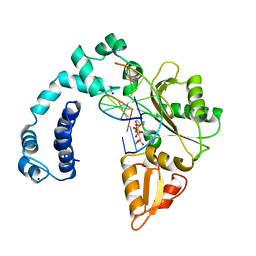 | |
3TIV
 
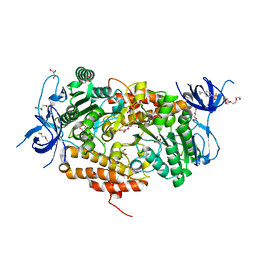 | | Crystal structure of subunit B mutant N157A of the A1AO ATP synthase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-08-22 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
8FE1
 
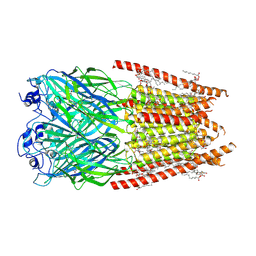 | | Alpha1/BetaB Heteromeric Glycine Receptor in 1 mM Glycine 20 uM Ivermectin State | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbs, E, Chakrapani, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor.
Nat Commun, 14, 2023
|
|
8ICZ
 
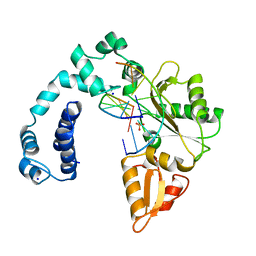 | | DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND LITHIUM SULFATE (75 MILLIMOLAR) | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*G)-3'), PROTEIN (DNA POLYMERASE BETA (E.C.2.7.7.7)), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1996-01-04 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis.
Biochemistry, 35, 1996
|
|
1BPZ
 
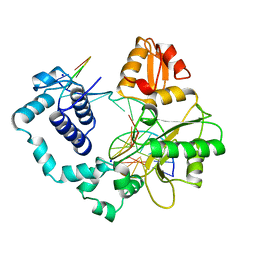 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Sawaya, M.R, Prasad, R, Wilson, S.H, Kraut, J, Pelletier, H. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
1BPY
 
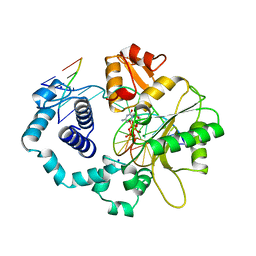 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*DOC)-3'), ... | | Authors: | Sawaya, M.R, Pelletier, H, Prasad, R, Wilson, S.H, Kraut, J. | | Deposit date: | 1997-04-15 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
5U0U
 
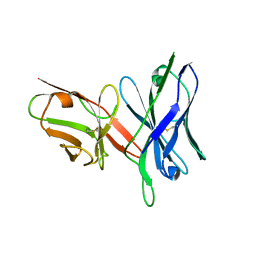 | |
1BPX
 
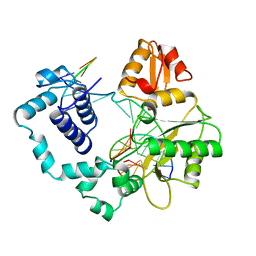 | | DNA POLYMERASE BETA/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Sawaya, M.R, Prasad, R, Wilson, S.H, Kraut, J, Pelletier, H. | | Deposit date: | 1997-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
5U15
 
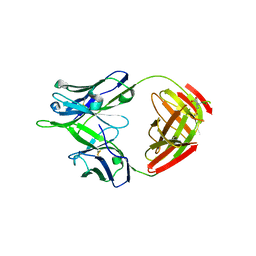 | |
5TQA
 
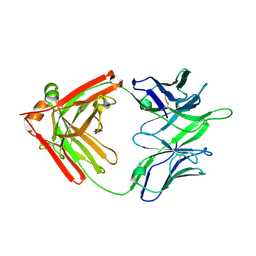 | |
5TRP
 
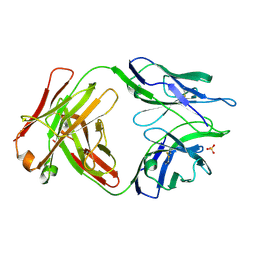 | |
5TPP
 
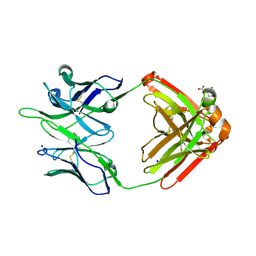 | |
5U0R
 
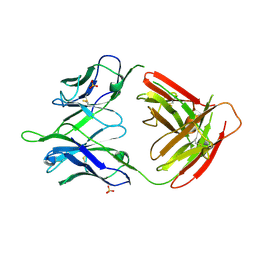 | |
5TPL
 
 | |
4YDV
 
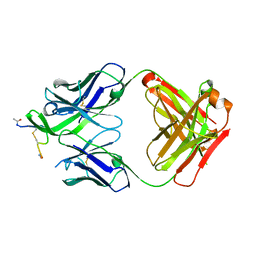 | | STRUCTURE OF THE ANTIBODY 7B2 THAT CAPTURES HIV-1 VIRIONS | | Descriptor: | HIV ANTIBODY 7B2 HEAVY CHAIN,IgG H chain, HIV ANTIBODY 7B2 LIGHT CHAIN,Ig kappa chain C region, HIV GP41 PEPTIDE GP41(596-606) | | Authors: | Nicely, N.I, Pemble IV, C.W. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques.
Plos Pathog., 11, 2015
|
|
8Z1S
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[4-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Amit, K, Swetha, R, Ghosh, K. | | Deposit date: | 2024-04-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atropisomerism Observed in Galactose-Based Monosaccharide Inhibitors of Galectin-3 Comprising 2-Methyl-4-phenyl-2,4-dihydro-3 H -1,2,4-triazole-3-thione.
J.Med.Chem., 67, 2024
|
|
8Z1T
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[4-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Amit, K, Swetha, R, Ghosh, K. | | Deposit date: | 2024-04-12 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atropisomerism Observed in Galactose-Based Monosaccharide Inhibitors of Galectin-3 Comprising 2-Methyl-4-phenyl-2,4-dihydro-3 H -1,2,4-triazole-3-thione.
J.Med.Chem., 67, 2024
|
|
