7CEE
 
 | | Crystal structure of mouse neuroligin-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-3 | | Authors: | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | Deposit date: | 2020-06-23 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
7CEG
 
 | | Crystal structure of the complex between mouse PTP delta and neuroligin-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform C of Receptor-type tyrosine-protein phosphatase delta, Neuroligin-3 | | Authors: | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | Deposit date: | 2020-06-23 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
3K38
 
 | |
3K39
 
 | |
3K36
 
 | |
3K37
 
 | | Crystal Structure of B/Perth Neuraminidase in complex with Peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Oakley, A.J, McKimm-Breschkin, J.L. | | Deposit date: | 2009-10-02 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Basis of Resistance to Neuraminidase Inhibitors of Influenza B Viruses.
J.Med.Chem., 2010
|
|
3K3A
 
 | |
3WDZ
 
 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | Descriptor: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
7E5P
 
 | | Aptamer enhancing peroxidase activity of myoglobin | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*AP*GP*GP*G)-3') | | Authors: | Tsukakoshi, K, Matsugami, A, Khunathai, K, Kanazashi, M, Yamagishi, Y, Nakama, K, Oshikawa, D, Hayashi, F, Kuno, H, Ikebukuro, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol.
Nucleic Acids Res., 49, 2021
|
|
3WV2
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydroquinazoline-2-carboxamide | | Descriptor: | CALCIUM ION, Collagenase 3, N-(3-methoxybenzyl)-4-oxo-3,4-dihydroquinazoline-2-carboxamide, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
3WV3
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
3WV1
 
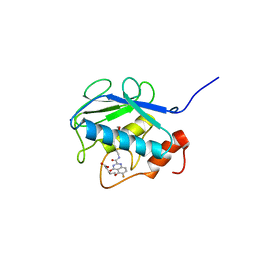 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-(2-((6-fluoro-2-((3-methoxybenzyl)carbamoyl)-4-oxo-3,4-dihydroquinazolin-5-yl)oxy)ethyl)benzoic acid | | Descriptor: | 4-[2-({6-fluoro-2-[(3-methoxybenzyl)carbamoyl]-4-oxo-3,4-dihydroquinazolin-5-yl}oxy)ethyl]benzoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of novel, highly potent, and selective quinazoline-2-carboxamide-based matrix metalloproteinase (MMP)-13 inhibitors without a zinc binding group using a structure-based design approach
J.Med.Chem., 57, 2014
|
|
2E8Z
 
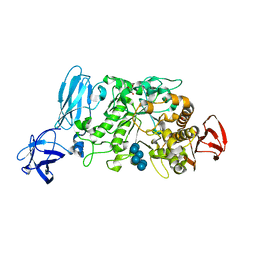 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with alpha-cyclodextrin | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E9B
 
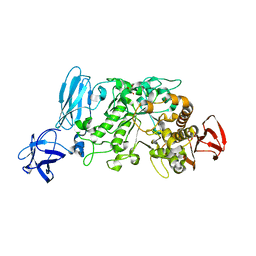 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with maltose | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E8Y
 
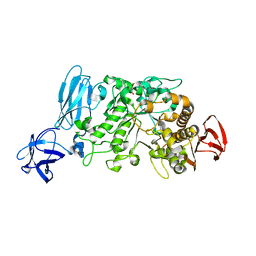 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168
in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2ZYZ
 
 | | Pyrobaculum aerophilum splicing endonuclease | | Descriptor: | Putative uncharacterized protein PAE0789, tRNA-splicing endonuclease | | Authors: | Yoshinari, S, Inaoka, D.K, Watanabe, Y, Shiba, T, Kurisu, G, Harada, S. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease
Nucleic Acids Res., 37, 2009
|
|
2WD5
 
 | | SMC hinge heterodimer (Mouse) | | Descriptor: | STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 1A, STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 3 | | Authors: | Michie, K.A, Haering, C.H, Nasmyth, K, Lowe, J. | | Deposit date: | 2009-03-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Channel within the Smc1/Smc3 Hinge Required for Sister Chromatid Cohesion.
Embo J., 30, 2011
|
|
2ZYC
 
 | | Crystal structure of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | PHOSPHATE ION, Peptidoglycan hydrolase FlgJ | | Authors: | Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2ZZR
 
 | |
