3F8T
 
 | |
7WCM
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist MBX-2982 | | Descriptor: | 2-[1-(5-ethylpyrimidin-2-yl)piperidin-4-yl]-4-[[4-(1,2,3,4-tetrazol-1-yl)phenoxy]methyl]-1,3-thiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
7WCN
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist AR231453 | | Descriptor: | Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
2MJV
 
 | |
2DHO
 
 | | Crystal structure of human IPP isomerase I in space group P212121 | | Descriptor: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-03-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
2JZY
 
 | |
7ES2
 
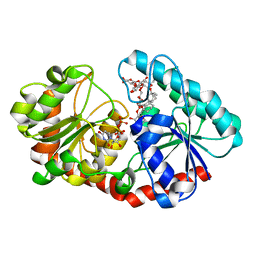 | | a mutant of glycosyktransferase in complex with UDP and Reb D | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, rebaudioside D | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERX
 
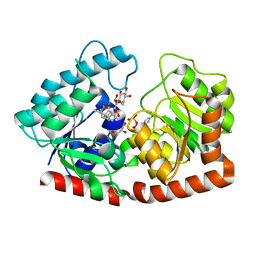 | | Glycosyltransferase in complex with UDP and STB | | Descriptor: | GLYCEROL, Glycosyltransferase, Steviolbioside, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES1
 
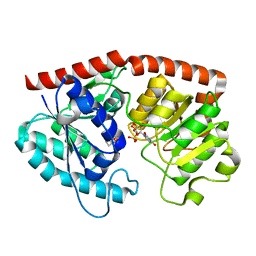 | | glycosyltransferase in complex with UDP and ST | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, steviol-19-o-glucoside | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES0
 
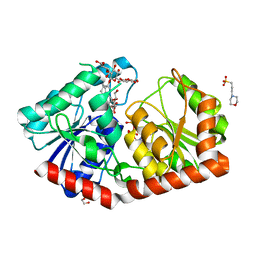 | | a rice glycosyltransferase in complex with UDP and REX | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERY
 
 | | apo form of the glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
2F90
 
 | | Crystal structure of bisphosphoglycerate mutase in complex with 3-phosphoglycerate and AlF4- | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, TETRAFLUOROALUMINATE ION | | Authors: | Wang, Y, Liu, L, Wei, Z, Gong, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase
J.Biol.Chem., 281, 2006
|
|
7V52
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V57
 
 | | Structure of AdaV | | Descriptor: | 2-OXOGLUTARIC ACID, AdaV, CHLORIDE ION, ... | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V54
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V56
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V7X
 
 | | Structure of H194A AdaV | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, AdaV | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7FIX
 
 | |
4FLN
 
 | | Crystal structure of plant protease Deg2 | | Descriptor: | Protease Do-like 2, chloroplastic, Unknown peptide | | Authors: | Gong, W, Liu, L, Sun, R, Gao, F. | | Deposit date: | 2012-06-15 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
J.Biol.Chem., 287, 2012
|
|
7FH5
 
 | | Structure of AdaV | | Descriptor: | AdaV, CHLORIDE ION, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
2WLU
 
 | | Iron-bound crystal structure of Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
