4IKG
 
 | |
4IDX
 
 | |
6AEJ
 
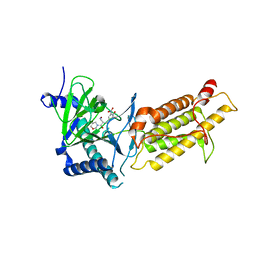 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (E)-3-[3-nitro-4,5-bis(oxidanyl)phenyl]-2-(1,3-oxazinan-3-ylcarbonyl)prop-2-enenitrile, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
4GS7
 
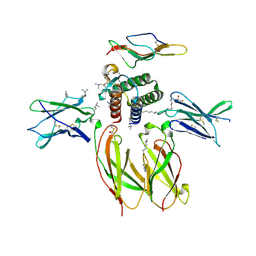 | | Structure of the Interleukin-15 quaternary complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ring, A.M, Ozkan, E, Feng, D, Garcia, K.C. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15.
Nat.Immunol., 13, 2012
|
|
6AK4
 
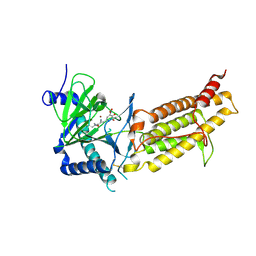 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (~{E})-2-cyano-~{N},~{N}-diethyl-3-[3-nitro-4,5-bis(oxidanyl)phenyl]prop-2-enamide, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
6K2D
 
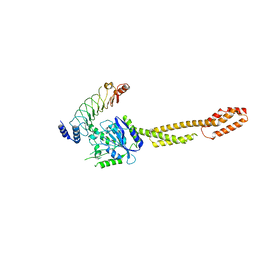 | | The crystal structure of GBP1 with LRR domain of IpaH9.8 | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8, Guanylate-binding protein 1 | | Authors: | Ji, C.G, Xiao, J.Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural mechanism for guanylate-binding proteins (GBPs) targeting by the Shigella E3 ligase IpaH9.8.
Plos Pathog., 15, 2019
|
|
6K1Z
 
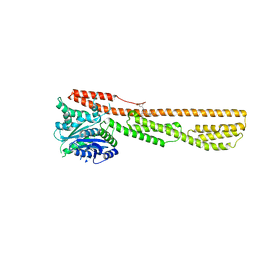 | | Crystal structure of farnesylated hGBP1 | | Descriptor: | FARNESYL, Guanylate-binding protein 1 | | Authors: | Du, S, Xiao, J.Y. | | Deposit date: | 2019-05-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structural mechanism for guanylate-binding proteins (GBPs) targeting by the Shigella E3 ligase IpaH9.8.
Plos Pathog., 15, 2019
|
|
6KN9
 
 | |
7UU6
 
 | |
7UUA
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase nsp5, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UU9
 
 | |
7UU8
 
 | |
7UUC
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UU7
 
 | |
7UUB
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UUE
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI85 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S,3R)-1-({(2S)-1-[2-(3-amino-3-oxopropyl)-2-propanoylhydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UUD
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
8XFB
 
 | | Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
8Z9M
 
 | | Cryo-EM structure of dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
7VPX
 
 | | The cryo-EM structure of the human pre-A complex | | Descriptor: | 5SS, DnaJ homolog subfamily C member 8, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-10-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
8DML
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt chenodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHENODEOXYCHOLIC ACID, ... | | Authors: | Tomchick, D.R, Orth, K, Zou, A.J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus.
J.Biol.Chem., 299, 2023
|
|
3OMZ
 
 | |
7KPR
 
 | |
7N0Z
 
 | |
7L4J
 
 | | Crystal structure of WT PPM1H phosphatase | | Descriptor: | MAGNESIUM ION, Protein phosphatase 1H | | Authors: | Khan, A.R, Waschbusch, D. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural basis for the specificity of PPM1H phosphatase for Rab GTPases.
Embo Rep., 22, 2021
|
|
