4FQH
 
 | | Crystal Structure of Fab CR9114 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, antibody CR9114 heavy chain, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FNK
 
 | |
7BYO
 
 | | Lysozyme structure SS1 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7BYP
 
 | | Lysozyme structure SASE1 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
8F92
 
 | | HIV Env BG505_MD39_B11 SOSIP boosting trimer in complex with B11_d77.7 mouse Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B11_d77.7 Fab heavy chain, ... | | Authors: | Torres, J.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | mRNA-LNP HIV-1 trimer boosters elicit precursors to broad neutralizing antibodies.
Science, 384, 2024
|
|
8F9G
 
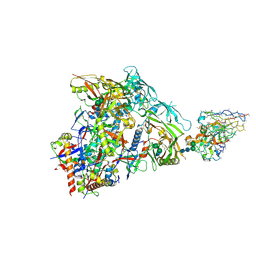 | | HIV Env germline targeting BG505_MD64_N332-GT5 SOSIP in complex with V3-glycan polyclonal Fab isolated from immunized BG18HCgl knock-in mice | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505_MD64_N332-GT5 gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | mRNA-LNP HIV-1 trimer boosters elicit precursors to broad neutralizing antibodies.
Science, 384, 2024
|
|
8F9M
 
 | |
8VFV
 
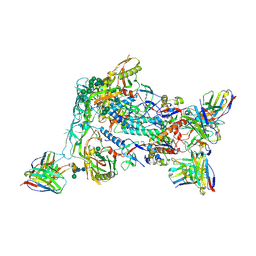 | | HIV Env BG505_MD39_B16 SOSIP boosting trimer in complex with B16_d77.5 mouse Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B16_d77.5 mouse Fab heavy chain Fv, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | mRNA-LNP HIV-1 trimer boosters elicit precursors to broad neutralizing antibodies.
Science, 384, 2024
|
|
6CGW
 
 | |
7D04
 
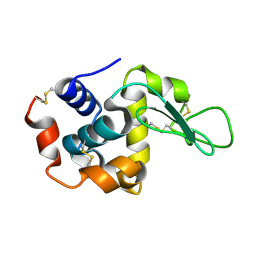 | | Lysozyme structure SS3 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D05
 
 | | Lysozyme structure SASE3 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D02
 
 | | Lysozyme structure SASE2 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D01
 
 | | Lysozyme structure SS2 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
6CHC
 
 | | JzTx-V toxin peptide, wild-type | | Descriptor: | Beta/kappa-theraphotoxin-Cg2a | | Authors: | Jordan, J.B. | | Deposit date: | 2018-02-22 | | Release date: | 2018-05-16 | | Method: | SOLUTION NMR | | Cite: | Pharmacological characterization of potent and selective NaV1.7 inhibitors engineered from Chilobrachys jingzhao tarantula venom peptide JzTx-V.
PLoS ONE, 13, 2018
|
|
8K41
 
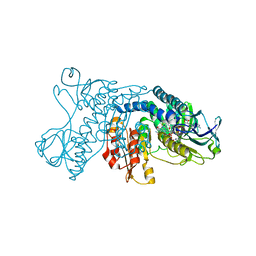 | | mercuric reductase,GbsMerA, - FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Do, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.
Sci Rep, 13, 2023
|
|
8K40
 
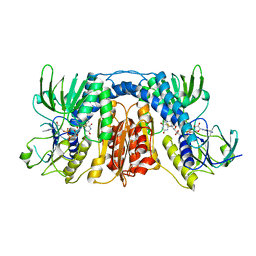 | | mercuric reductase,GbsMerA, - FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase | | Authors: | Do, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.
Sci Rep, 13, 2023
|
|
3OUR
 
 | |
7ERR
 
 | |
1WKO
 
 | |
1WKP
 
 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
5ZXU
 
 | |
5ZXN
 
 | | Crystal structure of CurA from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP-dependent oxidoreductase | | Authors: | Kim, M.-K, Bae, D.-W, Cha, S.-S. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Structural and Biochemical Characterization of the Curcumin-Reducing Activity of CurA from Vibrio vulnificus.
J. Agric. Food Chem., 66, 2018
|
|
4B18
 
 | |
5X9U
 
 | | Crystal structure of group III chaperonin in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome, alpha subunit | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
1JO7
 
 | | Solution Structure of Influenza A Virus Promoter | | Descriptor: | Influenza A virus promoter RNA | | Authors: | Bae, S.-H, Cheong, H.-K, Lee, J.-H, Cheong, C, Kainosho, M, Choi, B.-S. | | Deposit date: | 2001-07-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of an influenza virus promoter and their implications for viral RNA synthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
