1WB6
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with vanillate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4EK5
 
 | |
4EUM
 
 | | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II
To be Published
|
|
1YU4
 
 | | Major Tropism Determinant U1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-U1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
4EER
 
 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
2X0K
 
 | | Crystal structure of modular FAD synthetase from Corynebacterium ammoniagenes | | Descriptor: | PYROPHOSPHATE, RIBOFLAVIN BIOSYNTHESIS PROTEIN RIBF, SULFATE ION | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Medina, M, Frago, S. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligomeric State in the Crystal Structure of Modular Fad Synthetase Provides Insights Into its Sequential Catalysis in Prokaryotes
J.Mol.Biol., 400, 2010
|
|
1YOK
 
 | | crystal structure of human LRH-1 bound with TIF-2 peptide and phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, MacKay, J.A, Juzumiene, D, Bynum, J.M, Madauss, K, Montana, V, Lebedeva, L, Suzawa, M, Williams, J.D, Williams, S.P, Guy, R.K, Thornton, J.W, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1.
Cell(Cambridge,Mass.), 120, 2005
|
|
2WRT
 
 | | The 2.4 Angstrom structure of the Fasciola hepatica mu class GST, GST26 | | Descriptor: | CHLORIDE ION, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME 51 | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.5 Angstrom Structure of a Mu Class Gst from Fasciola Hepatica
To be Published
|
|
1YQV
 
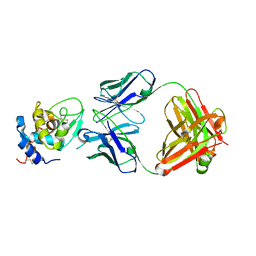 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4EK8
 
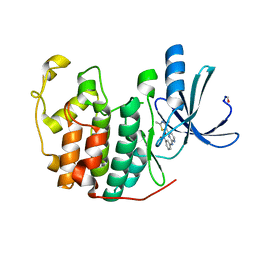 | |
2WMB
 
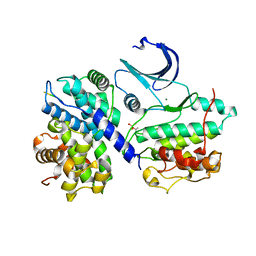 | | Structural and thermodynamic consequences of cyclization of peptide ligands for the recruitment site of cyclin A | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, LINEAR RKLFD, ... | | Authors: | Robertson, G.F, Endicott, J.A, Noble, M.E.M, McDonnell, J.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Consequences of Cyclization of Peptide Ligands for the Recruitment Site of Cyclin A
To be Published
|
|
2WK9
 
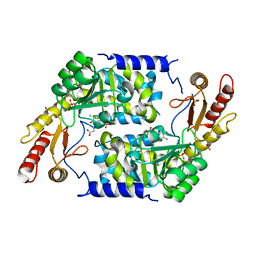 | | Structure of Plp_Thr aldimine form of Vibrio cholerae CqsA | | Descriptor: | CAI-1 AUTOINDUCER SYNTHASE, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Jahan, N, Potter, J.A, Sheikh, M.A, Botting, C.H, Shirran, S.L, Westwood, N.J, Taylor, G.L. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into the Biosynthesis of the Vibrio Cholerae Major Autoinducer Cai-1 from the Crystal Structure of the Plp-Dependent Enzyme Cqsa.
J.Mol.Biol., 392, 2009
|
|
152D
 
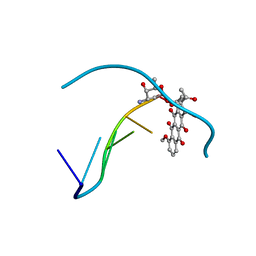 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
1YO2
 
 | | Proton Transfer from His200 in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Bhatt, D, Tu, C, Fisher, S.Z, Hernandez Prada, J.A, McKenna, R, Silverman, D.N. | | Deposit date: | 2005-01-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton transfer in a Thr200His mutant of human carbonic anhydrase II
Proteins, 61, 2005
|
|
4BXS
 
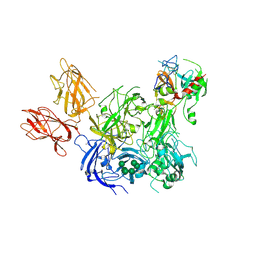 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
2W47
 
 | | Clostridium thermocellum CBM35 in complex with delta-4,5- anhydrogalacturonic acid | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, LIPOLYTIC ENZYME, ... | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W87
 
 | | Xyl-CBM35 in complex with glucuronic acid containing disaccharide. | | Descriptor: | CALCIUM ION, ESTERASE D, UNKNOWN LIGAND, ... | | Authors: | Montainer, C, Bueren, A.L.v, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1XU8
 
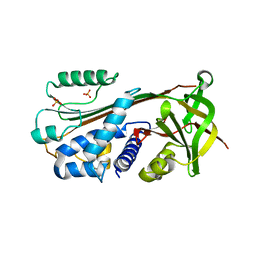 | | The 2.8 A structure of a tumour suppressing serpin | | Descriptor: | Maspin, SULFATE ION | | Authors: | Irving, J.A, Law, R.H, Ruzyla, K, Bashtannyk-Puhalovich, T.A, Kim, N, Worrall, D.M, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
2VTJ
 
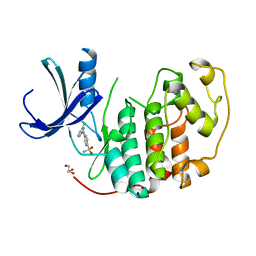 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 4-[(6-chloropyrazin-2-yl)amino]benzenesulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
4C76
 
 | |
2VTT
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-difluorophenyl)carbonyl]amino}-N-[(3S)-piperidin-3-yl]-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
1XL9
 
 | | Crystal Structure of Dihydrodipicolinate Synthase DapA-2 (BA3935) from Bacillus Anthracis. | | Descriptor: | dihydrodipicolinate synthase | | Authors: | Blagova, E, Levdikov, V, Milioti, N, Fogg, M.J, Kalliomaa, A.K, Brannigan, J.A, Wilson, K.S, Wilkinson, A.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase (BA3935) from Bacillus anthracis at 1.94 A resolution.
Proteins, 62, 2006
|
|
4BOL
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4CDH
 
 | | Crystallographic structure of the Human Igg1 alpha 2-6 sialilated Fc-Fragment | | Descriptor: | IG GAMMA-1 CHAIN C REGION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-11-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4BQP
 
 | | Mtb InhA complex with Methyl-thiazole compound 7 | | Descriptor: | (1S)-1-(5-{[1-(2,6-DIFLUOROBENZYL)-1H-PYRAZOL-3-YL]AMINO}-1,3,4-THIADIAZOL-2-YL)-1-(4-METHYL-1,3-THIAZOL-2-YL)ETHANOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
