8XS3
 
 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
8WQY
 
 | | Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WR0
 
 | | Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQV
 
 | | Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQU
 
 | | Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQX
 
 | | Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
1O63
 
 | |
1O6C
 
 | |
8GUS
 
 | | Cryo-EM structure of HU-CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUQ
 
 | | Cryo-EM structure of CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUT
 
 | | Cryo-EM structure of LEI-CB2-Gi complex | | Descriptor: | 1-[[4-[5-fluoranyl-6-[(oxan-4-ylamino)methyl]pyridin-2-yl]phenyl]methyl]-3-(2-methylpropyl)imidazolidine-2,4-dione, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, X.T, Chang, H, Wu, L.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUR
 
 | | Cryo-EM structure of CP-CB2-G protein complex | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
4O0U
 
 | |
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
1O66
 
 | |
1O60
 
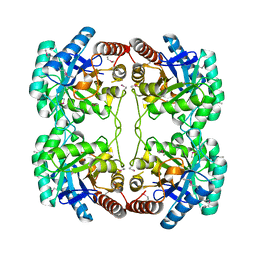 | | Crystal structure of KDO-8-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O65
 
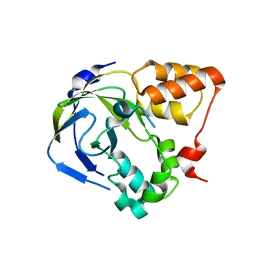 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O67
 
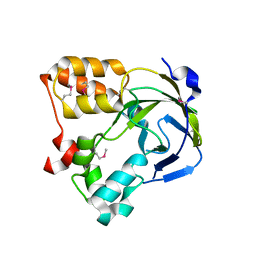 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O64
 
 | |
1O62
 
 | |
1O6D
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein TM0844 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O68
 
 | |
7WWH
 
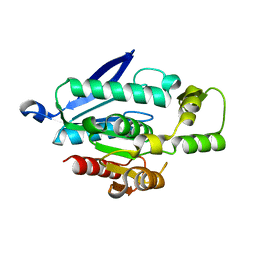 | |
