4IDV
 
 | | Crystal Structure of NIK with compound 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol (13V) | | Descriptor: | 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4EYT
 
 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 | | Descriptor: | SULFATE ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
5GNX
 
 | | The E171Q mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5GNZ
 
 | | The M3 mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
4DOU
 
 | | Crystal Structure of a Single-chain Trimer of Human Adiponectin Globular Domain | | Descriptor: | 1,2-ETHANEDIOL, Adiponectin, CALCIUM ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a single-chain trimer of human adiponectin globular domain.
Febs Lett., 586, 2012
|
|
5FDX
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
8X5I
 
 | | Structure of ATP/Mg2+ bound Gabija GajA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY4
 
 | | GajA tetramer with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X51
 
 | | Structure of DNA-bound GajA dimer (focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5N
 
 | | Structure of ATP/Mg2+ bound Gabija GajA-GajB 4:4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
4ERD
 
 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 in complex with stem IV of telomerase RNA | | Descriptor: | 5'-R(P*GP*GP*UP*CP*GP*AP*CP*AP*UP*CP*UP*UP*CP*GP*GP*AP*UP*GP*GP*AP*CP*C)-3', POTASSIUM ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
4LYC
 
 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
4LYB
 
 | | CdS within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
3TGV
 
 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
7JR7
 
 | | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8, Fab 11F4 heavy chain, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z, Zhang, H. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies
Commun Biol, 4, 2021
|
|
3TEF
 
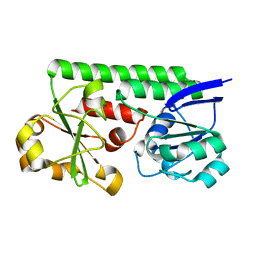 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
4PBD
 
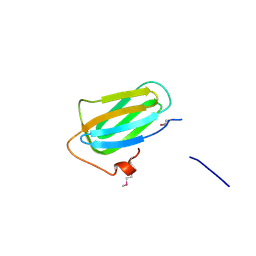 | |
4PCK
 
 | | Crystal structure of the P22S mutant of N-terminal CS domain of human Shq1 | | Descriptor: | GLYCEROL, Protein SHQ1 homolog | | Authors: | Singh, M, Wang, Z, Cascio, D, Feigon, J. | | Deposit date: | 2014-04-15 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure and Interactions of the CS Domain of Human H/ACA RNP Assembly Protein Shq1.
J.Mol.Biol., 427, 2015
|
|
7JTW
 
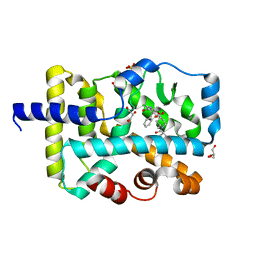 | | Crystal structure of RORgt with compound (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid | | Descriptor: | (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid, GLYCEROL, RAR-related orphan receptor C isoform a variant, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2020-08-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 6-Oxo-4-phenyl-hexanoic acid derivatives as ROR gamma t inverse agonists showing favorable ADME profile.
Bioorg.Med.Chem.Lett., 36, 2021
|
|
8WIG
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463S/Q484A | | Descriptor: | Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WII
 
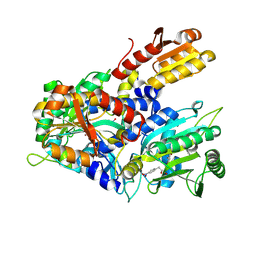 | | Crystal structure of E. coli ThrS catalytic domain mutant G463A in complex with Obafluorin | | Descriptor: | Threonine--tRNA ligase, ZINC ION, ~{N}-[(2~{R},3~{S})-2-[(4-nitrophenyl)methyl]-4-oxidanylidene-oxetan-3-yl]-2,3-bis(oxidanyl)benzamide | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WIJ
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant L489M in complex with Obafluorin | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WIA
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463S | | Descriptor: | Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WIH
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463A in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
